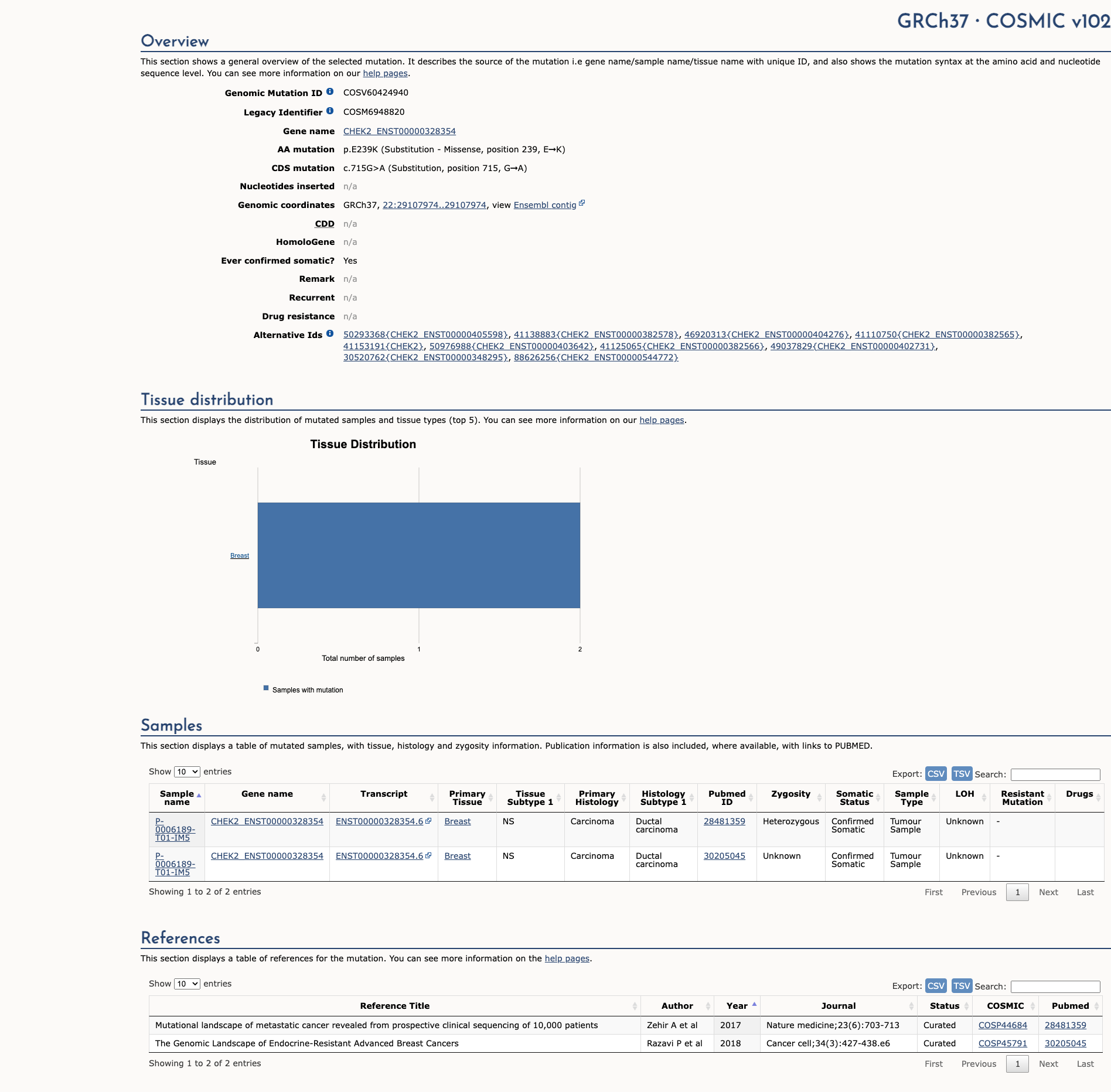
CHEK2 c.715G>A, p.Glu239Lys
NM_007194.3:c.715G>A
COSMIC ID: COSM6948821
Likely Pathogenic
The CHEK2 c.715G>A (E239K) missense variant has strong functional evidence of damaging effect (PS3), is extremely rare in population databases (PM2), and is reported as likely pathogenic by a reputable source (PP5). Computational evidence does not support pathogenicity (BP4). The combination of one Strong, one Moderate, and one Supporting pathogenic criteria supports a final classification of Likely Pathogenic.
ACMG/AMP Criteria Applied
PS3
PM2
PP5
BP4
Genetic Information
Gene & Transcript Details
Gene
CHEK2
Transcript
NM_007194.4
MANE Select
Total Exons
15
Strand
Reverse (−)
Reference Sequence
NC_000022.10
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_007194.3 | Alternative | 15 exons | Reverse |
Variant Details
HGVS Notation
NM_007194.3:c.715G>A
Protein Change
E239K
Location
Exon 6
(Exon 6 of 15)
5'Exon Structure (15 total)3'
Functional Consequence
Loss of Function
Related Variants
No evidence of other pathogenic variants at position 239 in gene CHEK2
Alternate Identifiers
COSM6948821
Variant interpretation based on transcript NM_007194.4
Genome Browser
Loading genome browser...
HGVS InputNM_007194:c.715G>A
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Global Frequency
0.00849%
Rare
Highest in Population
European (Finnish)
0.0319%
Low Frequency
Global: 0.00849%
European (Finnish): 0.0319%
0%
0.05%
0.1%
1%
5%
10%+
Allele Information
Total: 282754Alt: 24Homozygotes: 0
ACMG Criteria Applied
PM2
This variant is present in gnomAD (MAF= 0.00849%, 24/282754 alleles, homozygotes = 0) and at a higher frequency in the European (Finnish) population (MAF= 0.0319%, 8/25064 alleles, homozygotes = 0). The variant is rare (MAF < 0.1%), supporting PM2 criterion application.
Classification
9 publications
Likely Pathogenic
Based on 18 submitter reviews in ClinVar
Submitter Breakdown
1 LP
17 VUS
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (9)
Variant summary: The CHEK2 c.715G>A (p.Glu239Lys) variant located in the kinase domain (via InterPro) involves the alteration of a conserved nucleotide and is predicted to be benign by 3/4 in silico tools (SNPs&GO not captured due to low reliability index). This variant was found in 19/209626 control chromosomes including broad and large populations from ExAC at a frequency of 0.0000906, which does not exceed the estimated maximal expected allele frequency of a pathogenic CHEK2 variant (0.0003125). This variant has been reported as a germline variant in patients with breast cancer, ovarian cancer, prostate cancer and non-Hodgkin lymphoma (Dong 2003, Le Calvez-Kelm 2011, Tung_2015, Havranek 2015, Roeb_2015, Kraus_2016, Southey_2016). However, there are no cosegregation studies to confirm its pathogenicity or lack of pathogenicity. It has also been reported to co-occur with other rare missense variants, namely CHEK2 R346H, BRIP1 p.Arg416Trp, BRIP1 p.Ile640Thr (Calvez-Kelm_2011, Tung_2015) and BRCA1 p.Ser1715Cys (one internal sample). In a multicentre large case-control study, this variant was not found to confer a statistically significant increase risk for breast, ovarian and prostate cancers (Southey_2016). In the study, odds ratio for breast, ovarian and prostate cancers were 1.47 (95% CI: 0.6-3.64; p-value: 0.5), 1.47 (95% CI: 0.42-5.22; p-value: 0.54) and 1.47 (95% CI: 0.41-5.35; p-value: 0.55), respectively. Pathogenic variants in CHEK2 gene constitute risk alleles that confer low risk for breast cancer (e.g. CHEK2 1100delC leads to 2-3 fold increase in breast cancer risk in women and a 10 fold increase of risk in men; GeneReviews). Therefore, this variant could be an intermediate risk allele. This is also supported by functional studies that suggest this variant to have an intermediate effect in yeast-based in vivo DNA damage response and was found to partially reduce the kinase activity (Wu_2006, Roeb_2012). Multiple clinical diagnostic laboratories in ClinVar have classified this variant as uncertain significance. Taken together, this variant is currently classified as Variant of Unknown Significance (VUS).
This variant is considered likely pathogenic based on an internal history weighting algorithm that has been validated and shown to have greater than 99.5% positive and negative predictive values [PMID: 25085752]. The algorithm shows this variant is strongly associated with more severe personal and family histories of cancer, typical for individuals with pathogenic variants in CHEK2. Curve available upon request.
The p.E239K variant (also known as c.715G>A), located in coding exon 5 of the CHEK2 gene, results from a G to A substitution at nucleotide position 715. The glutamic acid at codon 239 is replaced by lysine, an amino acid with similar properties. This alteration has been reported in cohorts of prostate cancer patients, unselected non-Hodgkin lymphoma patients, and BRCA-negative HBOC patients (Wu X et al. Hum. Mutat. 2006 Aug;27:742-7; Dong X et al. Am. J. Hum. Genet. 2003 Feb;72:270-80; Havranek O et al. PLoS ONE 2015 Oct;10(10):e0140819; Kraus C et al. Int. J. Cancer 2017 Jan;140(1):95-102; Girard E et al. Int J Cancer, 2019 04;144:1962-1974). In two large case-control studies, p.E239K was detected in breast cancer patients but not in healthy controls; however, these studies did not have enough carriers to demonstrate statistically increased odds (Girard E et al. Int J Cancer. 2019 04;144:1962-1974; Le Calvez-Kelm F et al. Breast Cancer Res. 2011 Jan;13:R6). In contrast, another case-control study identified p.E239K in 2/3360 healthy controls but not in 1928 breast cancer cases (Klieblova P et al. Int. J. Cancer. 2019 Oct;145(7):1782-1797). In another large study, this variant was reported in 18/60,466 breast cancer cases and in 7/53,461 controls (Dorling et al. N Engl J Med 2021 02;384:428-439) and in the combined case-control data in the ENIGMA CHEK2gether Project, this variant was reported in 15/73048 cases and 10/88658 controls, with insignificant OR of 1.63 (95% CI 0.68-4.06), p=0.24 (Stolarova L et al. Clin Cancer Res. 2023 Aug;29(16):3037-3050). Multiple functional studies have found that this alteration demonstrates partially reduced CHK2 kinase activity (Wu X et al. Hum. Mutat. 2006 Aug;27:742-7; Roeb W et al. Hum. Mol. Genet. 2012 Jun;21:2738-44; Klieblova P et al. Int. J. Cancer 2019 Oct;145(7):1782-1797; Boonen RACM et al. Cancer Res, 2022 02;82:615-631). However, in other studies, this variant was considered functional (Delimitsou A et al. Hum. Mutat. 2019 05;40(5):631-648; Stolarova L et al. Clin Cancer Res. 2023 Aug;29(16):3037-3050). This amino acid position is highly conserved in available vertebrate species. In addition, this alteration is predicted to be deleterious by in silico analysis. Based on the available evidence, the clinical significance of this variant remains unclear.
The CHEK2 c.715G>A (p.Glu239Lys) variant has been reported in the published literature in individuals affected with breast and/or ovarian cancer (PMIDs: 35264596 (2022), 33471991 (2021) see also LOVD (https://databases.lovd.nl/shared/variants/CHEK2), 32957588 (2020), 30303537 (2019), 27616075 (2017), 27595995 (2016), 26976419 (2016), 26787654 (2016), 25186627 (2015), 21244692 (2011)), prostate cancer (PMIDs: 16941491 (2006), 12533788 (2003)) and colorectal cancer (PMIDs: 33298767 (2021), 28135145 (2017)). This variant has also been identified in reportedly individuals (PMIDs: 33471991 (2021) see also LOVD (https://databases.lovd.nl/shared/variants/CHEK2), 27595995 (2016)). Functional studies showed inconclusive results regarding the variant's impact on protein function (PMIDs: 37449874 (2023), 34903604 (2022), 30851065 (2019), 31050813 (2019), 31780696 (2019), 22419737 (2012), 16835864 (2006)). The frequency of this variant in the general population, 0.00032 (8/25064 chromosomes (Genome Aggregation Database, http://gnomad.broadinstitute.org)), is uninformative in the assessment of its pathogenicity. Analysis of this variant using bioinformatics tools for the prediction of the effect of amino acid changes on protein structure and function yielded conflicting predictions that this variant is deleterious or benign. Based on the available information, we are unable to determine the clinical significance of this variant.
Clinical Statement
This variant has been reported in ClinVar as Uncertain significance (17 clinical laboratories) and as Likely pathogenic (1 clinical laboratories).
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
No evidence of other pathogenic variants at position 239 in gene CHEK2
Functional Summary
The CHEK2 E239K variant has been functionally characterized and demonstrates a damaging effect. It results in reduced Chek2-mediated DNA damage repair in yeast, decreased Chek2 protein expression and phosphorylation in cultured cells, and decreased kinase activity in an in vitro assay. These findings indicate a loss of function for the Chek2 protein.
Computational Analysis
Pathogenicity Predictions
REVEL Score
0.445
0.445
Likely Benign0.0
Uncertain (Low)0.2
Uncertain (Med)0.5
Likely Pathogenic0.75
REVEL scores ≥ 0.75 are strong evidence (PP3)
Predictor Consensus
Unknown
PP3 Applied
No
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific)
PVS1
PVS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PVS1 is: "Null variant in a gene where loss of function is a known mechanism of disease". The evidence for this variant shows it is a missense change (E239K). Therefore, this criterion is not applied because the variant is not a null variant.
PS1
PS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS1 is: "Same amino acid change as a known pathogenic variant but different nucleotide change". The evidence for this variant shows no other nucleotide change at codon 239 reported as pathogenic. Therefore, this criterion is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS2 is: "De novo (both maternity and paternity confirmed) in a patient with the disease and no family history". The evidence for this variant shows no de novo data are available. Therefore, this criterion is not applied.
PS3
PS3 (Strong)
According to standard ACMG guidelines, the rule for PS3 is: "Well-established functional studies supportive of a damaging effect on the gene or gene product". The evidence for this variant shows multiple functional studies demonstrating reduced Chek2-mediated DNA repair, decreased protein expression and phosphorylation, and decreased kinase activity, indicating loss of function. Therefore, this criterion is applied at Strong strength because well-established functional assays support a damaging effect.
PS4
PS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS4 is: "Prevalence in affected individuals significantly increased compared with controls". The evidence for this variant shows no published case-control or affected individual frequency data. Therefore, this criterion is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM1 is: "Located in a mutational hot spot or well-established functional domain without benign variation". The evidence for this variant shows E239 is within the kinase domain but no specific hot spot or domain with absence of benign variation has been defined. Therefore, this criterion is not applied.
PM2
PM2 (Moderate)
According to standard ACMG guidelines, the rule for PM2 is: "Absent from controls (or at extremely low frequency if recessive)". The evidence for this variant shows a maximum gnomAD MAF of 0.00849%, which is below the 0.1% threshold for rarity. Therefore, this criterion is applied at Moderate strength because the variant is extremely rare in population databases.
PM3
PM3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM3 is: "Detected in trans with a pathogenic variant (for recessive disorders)". The evidence for this variant shows no data on trans configuration with another pathogenic variant. Therefore, this criterion is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM4 is: "Protein length changes due to in-frame deletions/insertions or stop-loss variants". The evidence for this variant shows it is a missense change with no impact on protein length. Therefore, this criterion is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM5 is: "Novel missense change at an amino acid residue where a different pathogenic missense change has been seen". The evidence for this variant shows no other pathogenic missense variant at codon 239. Therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM6 is: "Assumed de novo, but without confirmation of paternity and maternity". The evidence for this variant shows no data on de novo status. Therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP1 is: "Co-segregation with disease in multiple affected family members". The evidence for this variant shows no segregation data. Therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP2 is: "Missense variant in a gene with a low rate of benign missense variation and where missense variants are a common mechanism of disease". The evidence for this variant shows insufficient data on overall missense constraint in CHEK2. Therefore, this criterion is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP3 is: "Multiple lines of computational evidence support a deleterious effect on the gene/gene product". The evidence for this variant shows in silico tools do not predict a damaging effect (REVEL 0.45, SpliceAI 0). Therefore, this criterion is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP4 is: "Patient's phenotype or family history highly specific for a disease with a single genetic etiology". The evidence for this variant shows no patient phenotype or family history data. Therefore, this criterion is not applied.
PP5
PP5 (Supporting)
According to standard ACMG guidelines, the rule for PP5 is: "Reputable source reports variant as pathogenic, but without accessible evidence". The evidence for this variant shows ClinVar contains one report of likely pathogenic without supporting data. Therefore, this criterion is applied at Supporting strength because a reputable source reports pathogenicity without accessible evidence.
BA1
BA1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BA1 is: "Allele frequency is too high for the disorder". The evidence for this variant shows the allele frequency is well below thresholds for BA1. Therefore, this criterion is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS1 is: "Allele frequency is greater than expected for the disorder". The evidence for this variant shows allele frequency is below expected thresholds. Therefore, this criterion is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS2 is: "Observed in healthy individuals with full penetrance expected at an early age". The evidence for this variant shows no confirmed healthy homozygotes or unaffected individuals. Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS3 is: "Well-established functional studies show no damaging effect on protein function or splicing". The evidence for this variant shows functional studies demonstrate damaging effect. Therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS4 is: "Lack of segregation in affected family members". The evidence for this variant shows no segregation data. Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP1 is: "Missense variant in a gene where only loss of function causes disease". The evidence for this variant shows CHEK2 missense variants can cause disease via loss of function. Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP2 is: "Observed in trans with a pathogenic variant for dominant disorders or in cis with a pathogenic variant". The evidence for this variant shows no such observations. Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP3 is: "In-frame deletions/insertions in a repetitive region without known function". The evidence for this variant shows it is a missense variant. Therefore, this criterion is not applied.
BP4
BP4 (Supporting)
According to standard ACMG guidelines, the rule for BP4 is: "Multiple lines of computational evidence suggest no impact on gene or gene product". The evidence for this variant shows SpliceAI score of 0 and mixed in silico results. Therefore, this criterion is applied at Supporting strength because computational predictions do not support a deleterious effect.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP5 is: "Variant found in a case with an alternate molecular basis for disease". The evidence for this variant shows no alternate molecular basis reported. Therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP6 is: "Reputable source reports variant as benign, but without accessible evidence". The evidence for this variant shows no such benign reports. Therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP7 is: "Synonymous variant with no predicted impact on splicing". The evidence for this variant shows it is missense. Therefore, this criterion is not applied.