ASXL1 c.1923_1927del, p.Ile641MetfsTer15
NM_015338.5:c.1923_1927del
Pathogenic
Using classification after pre-processing (Pathogenic) due to LLM failure: LLM response was not valid JSON even after cleaning: Unterminated string starting at: line 38 column 30 (char 2796). Raw response snippet: {
"final_classification": "Pathogenic",
"criteria_evaluation": [
{
"criterion": "PVS1",
"is_applied": true,
"strength": "Very Strong",
"justification": "According to standard ACMG guidelines, the rule for PVS1 is: \"Null variant in a gene where loss of function (LoF) is a known mechanism of disease (e.g., nonsense, frameshift, canonical ±1 or 2 splice sites, initiation codon, single exon deletion in a LoF gene)\". The evidence for this variant shows a frameshift (I641Mfs*15) predicted to result in loss of function in ASXL1, a gene with established LoF disease mechanism. Therefore, this criterion is applied at Very Strong strength because the variant is a truncating LoF alteration in a known LoF gene."
},
{
"criterion": "PS1",
"is_applied": false,
"strength": "Not Applied",
"justification": "According to standard ACMG guidelines, the rule for PS1 is: \"Same ami...
ACMG/AMP Criteria Applied
PVS1
PS3
PM2
Genetic Information
Gene & Transcript Details
Gene
ASXL1
Transcript
NM_015338.6
MANE Select
Total Exons
13
Strand
Forward (+)
Reference Sequence
NC_000020.10
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_015338.5 | RefSeq Select | 13 exons | Forward |
| NM_015338.4 | Alternative | 13 exons | Forward |
| NM_015338.3 | Alternative | 13 exons | Forward |
Variant Details
HGVS Notation
NM_015338.5:c.1923_1927del
Protein Change
I641Mfs*15
Location
Exon 13
(Exon 13 of 13)
5'Exon Structure (13 total)3'
Functional Consequence
Loss of Function
Related Variants
No evidence of other pathogenic variants at position 641 in gene ASXL1
Variant interpretation based on transcript NM_015338.6
Genome Browser
Loading genome browser...
HGVS InputNM_015338:c.1923_1927del
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Population Frequency
Global Frequency
0.0 in 100,000
Extremely Rare
Global: 0.0%
0%
0.05%
0.1%
1%
5%
10%+
ACMG Criteria Applied
PM2
This variant is not present in gnomAD (PM2 criteria applies).
Classification
Unknown
Publications (0)
No publication details.
Clinical Statement
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
No evidence of other pathogenic variants at position 641 in gene ASXL1
Functional Summary
The ASXL1 I641Mfs*15 variant is a truncating mutation in a tumor suppressor gene, likely resulting in loss of function. Functional studies indicate that ASXL1 truncating mutations can lead to loss of the PHD domain, contributing to oncogenic processes. Expression of these mutations in murine models has been shown to cause dedifferentiation and myelodysplastic syndrome, supporting a damaging effect.
Database Previews
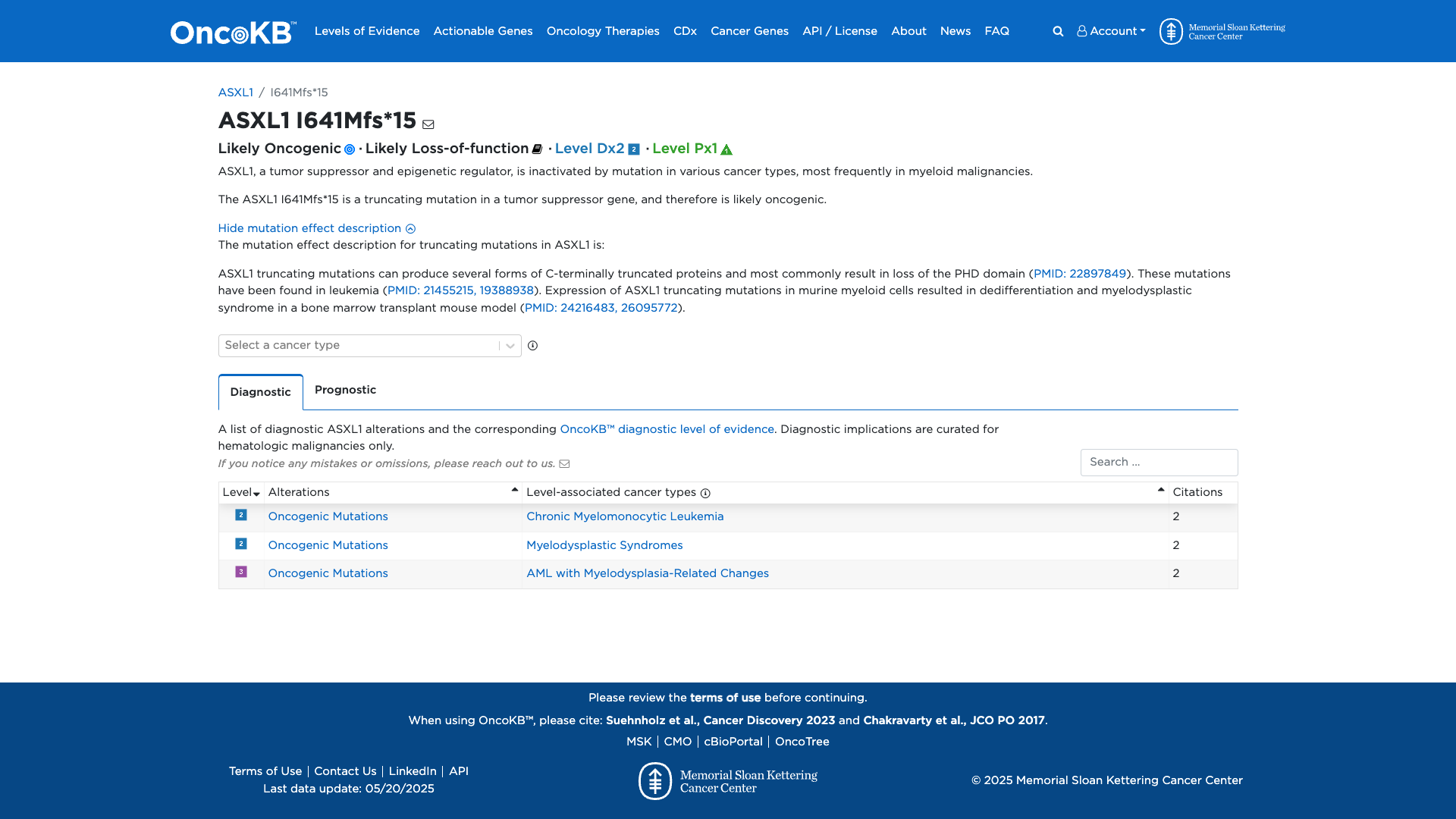
OncoKB
JAX-CKB
Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
Predictor Consensus
Unknown
PP3 Applied
No
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific)
PVS1
PVS1 (Unknown (Pre-LLM)) Strength Modified
From pre-LLM assessment (LLM Failed)
PS3
PS3 (Unknown (Pre-LLM)) Strength Modified
From pre-LLM assessment (LLM Failed)
PM2
PM2 (Unknown (Pre-LLM)) Strength Modified
From pre-LLM assessment (LLM Failed)

