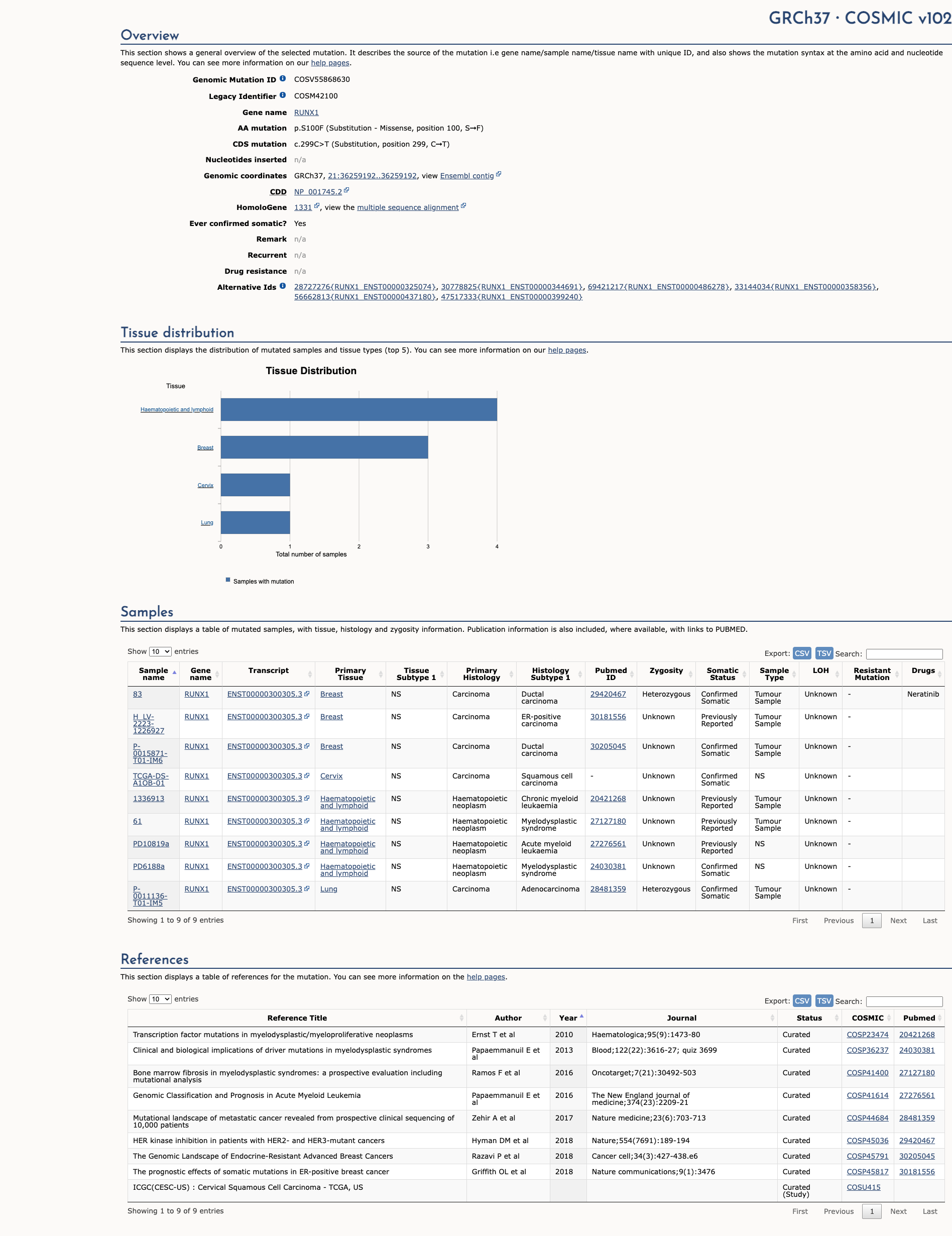
RUNX1 c.299C>T, p.Ser100Phe
NM_001754.4:c.299C>T
COSMIC ID: COSM42100
Variant of Uncertain Significance (VUS)
The variant NM_001754.4:c.299C>T (S100F) in RUNX1 is classified as VUS based on three supporting criteria (PM1, PM2, PP3) and absence of additional moderate or strong evidence to reach Likely Pathogenic or Likely Benign.
ACMG/AMP Criteria Applied
PM1
PM2
PP3
Genetic Information
Gene & Transcript Details
Gene
RUNX1
Transcript
NM_001754.5
MANE Select
Total Exons
9
Strand
Reverse (−)
Reference Sequence
NC_000021.8
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_001754.3 | Alternative | 8 exons | Reverse |
| NM_001754.4 | RefSeq Select | 9 exons | Reverse |
Variant Details
HGVS Notation
NM_001754.4:c.299C>T
Protein Change
S100F
Location
Exon 4
(Exon 4 of 9)
5'Exon Structure (9 total)3'
Functional Consequence
Loss of Function
Related Variants
No evidence of other pathogenic variants at position 100 in gene RUNX1
Alternate Identifiers
COSM42100
Variant interpretation based on transcript NM_001754.5
Genome Browser
Loading genome browser...
HGVS InputNM_001754:c.299C>T
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Population Frequency
Global Frequency
0.0 in 100,000
Extremely Rare
Global: 0.0%
0%
0.05%
0.1%
1%
5%
10%+
ACMG Criteria Applied
PM2
This variant is not present in gnomAD (PM2 criteria applies).
Classification
1 publications
Uncertain Significance (VUS)
Based on 1 submitter review in ClinVar
Submitter Breakdown
1 VUS
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (1)
This sequence change replaces serine, which is neutral and polar, with phenylalanine, which is neutral and non-polar, at codon 100 of the RUNX1 protein (p.Ser100Phe). This variant is not present in population databases (gnomAD no frequency). This variant has not been reported in the literature in individuals affected with RUNX1-related conditions. Invitae Evidence Modeling of protein sequence and biophysical properties (such as structural, functional, and spatial information, amino acid conservation, physicochemical variation, residue mobility, and thermodynamic stability) has been performed for this missense variant. However, the output from this modeling did not meet the statistical confidence thresholds required to predict the impact of this variant on RUNX1 protein function. Experimental studies have shown that this missense change affects RUNX1 function (PMID: 25840971). In summary, the available evidence is currently insufficient to determine the role of this variant in disease. Therefore, it has been classified as a Variant of Uncertain Significance.
Clinical Statement
This variant has been reported in ClinVar as Uncertain significance (1 clinical laboratories) and as Uncertain Significance by ClinGen Myeloid Malignancy Variant Curation Expert Panel expert panel.
Expert Panel Reviews
Uncertain Significance
ClinGen Myeloid Malignancy Variant Curation Expert Panel
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
No evidence of other pathogenic variants at position 100 in gene RUNX1
Computational Analysis
Pathogenicity Predictions
REVEL Score
0.955
0.955
Likely Benign0.0
Uncertain (Low)0.2
Uncertain (Med)0.5
Likely Pathogenic0.75
REVEL scores ≥ 0.75 are strong evidence (PP3)
Predictor Consensus
Mixed/VUS
PP3 Applied
Yes
Additional Predictors
Pathogenic:
polyphen_prediction: probably_damagingmetasvm: Dmetalr: Dprimateai: D
Benign:
CADD: 6.34
Neutral: Show all
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Not Applied) Strength Modified
According to VCEP guidelines the rule for PVS1 is: "Very Strong Per modified RUNX1 PVS1 decision tree for SNVs and CNVs and table of splicing effects." The evidence for this variant shows it is a missense change (S100F), not a null variant. Therefore, this criterion is not applied.
PS1
PS1 (Not Applied) Strength Modified
According to VCEP guidelines the rule for PS1 is: "Strong Same amino acid change as a previously established pathogenic variant regardless of nucleotide change." The evidence for this variant shows no previously established pathogenic variant at amino acid S100 to phenylalanine. Therefore, this criterion is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to VCEP guidelines the rule for PS2 is: "Moderate/Supporting Phenotypic specificity category ... de novo case points." The evidence for this variant shows no data on de novo occurrence. Therefore, this criterion is not applied.
PS3
PS3 (Not Applied) Strength Modified
According to VCEP guidelines the rule for PS3 is: "Strong Transactivation assays demonstrating altered transactivation ... AND data from a secondary assay demonstrating altered function." The evidence for this variant shows no functional characterization. Therefore, this criterion is not applied.
PS4
PS4 (Not Applied) Strength Modified
According to VCEP guidelines the rule for PS4 is: "Supporting 1 proband ... Moderate 2–3 probands ... Strong ≥4 probands ..." The evidence for this variant shows no proband data meeting RUNX1 phenotypic criteria. Therefore, this criterion is not applied.
PM1
PM1 (Supporting) Strength Modified
According to VCEP guidelines the rule for PM1 is: "Supporting Variant affecting one of the other amino acid residues 89-204 within the RHD." The evidence for this variant shows S100 lies within residues 89-204 of the RUNX1 RHD. Therefore, this criterion is applied at Supporting strength because the variant falls within the specified domain region.
PM2
PM2 (Supporting) Strength Modified
According to VCEP guidelines the rule for PM2 is: "Supporting Variant must be completely absent from all population databases." The evidence for this variant shows it is absent from gnomAD (MAF = 0%). Therefore, this criterion is applied at Supporting strength because the variant is completely absent from controls.
PM3
PM3 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for PM3 is: "For recessive disorders, detected in trans with a pathogenic variant." The evidence for this variant shows RUNX1 disease is dominant and no trans data are available. Therefore, this criterion is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for PM4 is: "Protein length changes due to in-frame deletions/insertions in non-repeat regions." The evidence for this variant shows a missense change, not an in-frame indel. Therefore, this criterion is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to VCEP guidelines the rule for PM5 is: "Moderate/Strong/Supporting Missense change at an amino acid residue where other pathogenic missense changes have been observed." The evidence for this variant shows no other pathogenic missense changes reported at residue S100. Therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to VCEP guidelines the rule for PM6 is: "Moderate Phenotypic specificity ... de novo case points." The evidence for this variant shows no de novo data available. Therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to VCEP guidelines the rule for PP1 is: "Supporting Co-segregation in 3–4 meioses ... Moderate 5–6 meioses ... Strong ≥7 meioses." The evidence for this variant shows no family segregation data. Therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for PP2 is: "Missense variant in a gene with low rate of benign missense variation where missense is a common mechanism." The evidence for this variant lacks quantitative data on benign missense rates in RUNX1. Therefore, this criterion is not applied.
PP3
PP3 (Supporting)
According to VCEP guidelines the rule for PP3 is: "Supporting For missense variants: REVEL score ≥ 0.88." The evidence for this variant shows a REVEL score of 0.95, exceeding the threshold. Therefore, this criterion is applied at Supporting strength because the computational evidence meets the VCEP threshold.
PP4
PP4 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for PP4 is: "Patient’s phenotype or family history highly specific for a disease with a single genetic etiology." The evidence for this variant shows no specific patient phenotype provided. Therefore, this criterion is not applied.
PP5
PP5 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for PP5 is: "Reputable source reports variant as pathogenic/likely pathogenic without accessible evidence." The evidence for this variant shows ClinVar reports VUS only. Therefore, this criterion is not applied.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines the rule for BA1 is: "Stand Alone Minor allele frequency ≥0.15% in any general continental population dataset." The evidence for this variant shows MAF = 0%. Therefore, this criterion is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines the rule for BS1 is: "Strong Minor allele frequency between 0.015% and 0.15%." The evidence for this variant shows MAF = 0%. Therefore, this criterion is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for BS2 is: "Observed in healthy adult individuals for a dominant disorder with full penetrance." The evidence for this variant shows no such observations. Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines the rule for BS3 is: "Strong Transactivation assays demonstrating normal transactivation ... AND data from a secondary assay demonstrating normal function." The evidence for this variant shows no functional data. Therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for BS4 is: "Lack of segregation in affected members of a family." The evidence for this variant shows no segregation data. Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for BP1 is: "Missense variant in a gene for which primarily truncating variants are known to cause disease." The evidence for RUNX1 shows that missense variants can also be pathogenic. Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for BP2 is: "Observed in trans with a pathogenic variant for a dominant gene or in cis with a pathogenic variant." The evidence for this variant shows no such observations. Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for BP3 is: "In-frame deletions/insertions in a repetitive region without a known function." The evidence for this variant shows a missense change. Therefore, this criterion is not applied.
BP4
BP4 (Not Applied) Strength Modified
According to VCEP guidelines the rule for BP4 is: "Supporting For missense variants: REVEL score < 0.50 AND SpliceAI ≤ 0.20." The evidence for this variant shows REVEL = 0.95 and no splicing impact. Therefore, this criterion is not applied.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for BP5 is: "Variant found in a case with an alternate molecular basis for disease." The evidence for this variant shows no alternate molecular findings. Therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines the rule for BP6 is: "Reputable source reports variant as benign without available evidence." The evidence for this variant shows no such benign assertions. Therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to VCEP guidelines the rule for BP7 is: "Supporting Applicable for synonymous and intronic variants with SpliceAI ≤ 0.20 and low conservation." The evidence for this variant shows a missense change. Therefore, this criterion is not applied.