TP53 c.877G>T, p.Gly293Trp
NM_000546.6:c.877G>T
COSMIC ID: COSM46261
Likely Pathogenic
Under TP53 VCEP guidelines, only PM2_Supporting is met (rare in population). No other criteria meet VCEP specifications due to lack of validated functional assay data or absence of hotspot, segregation, or computational evidence. Classification is VUS.
ACMG/AMP Criteria Applied
PM2
Genetic Information
Gene & Transcript Details
Gene
TP53
Transcript
NM_000546.6
MANE Select
Total Exons
11
Strand
Reverse (−)
Reference Sequence
NC_000017.10
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_000546.5 | RefSeq Select | 11 exons | Reverse |
| NM_000546.3 | Alternative | 11 exons | Reverse |
| NM_000546.4 | Alternative | 11 exons | Reverse |
| NM_000546.2 | Alternative | 11 exons | Reverse |
Variant Details
HGVS Notation
NM_000546.6:c.877G>T
Protein Change
G293W
Location
Exon 8
(Exon 8 of 11)
5'Exon Structure (11 total)3'
Functional Consequence
Loss of Function
Related Variants
No evidence of other pathogenic variants at position 293 in gene TP53
Alternate Identifiers
COSM46261
Variant interpretation based on transcript NM_000546.6
Genome Browser
Loading genome browser...
HGVS InputNM_000546:c.877G>T
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Global Frequency
0.00106%
Rare
Highest in Population
European (non-Finnish)
0.00232%
Rare
Global: 0.00106%
European (non-Finnish): 0.00232%
0%
0.05%
0.1%
1%
5%
10%+
Allele Information
Total: 282876Alt: 3Homozygotes: 0
ACMG Criteria Applied
PM2
This variant is present in gnomAD (MAF= 0.00106%, 3/282876 alleles, homozygotes = 0) and at a higher frequency in the European (non-Finnish) population (MAF= 0.00232%, 3/129184 alleles, homozygotes = 0). The variant is rare (MAF < 0.1%), supporting PM2 criterion application.
Classification
7 publications
Uncertain Significance (VUS)
Based on 8 submitter reviews in ClinVar
Submitter Breakdown
5 VUS
3 LB
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (7)
This alteration is classified as likely benign based on a combination of the following: seen in unaffected individuals, population frequency, intact protein function, lack of segregation with disease, co-occurrence, RNA analysis, in silico models, amino acid conservation, lack of disease association in case-control studies, and/or the mechanism of disease or impacted region is inconsistent with a known cause of pathogenicity.
Variant summary: TP53 c.877G>T (p.Gly293Trp) results in a non-conservative amino acid change in the encoded protein sequence. Three of five in-silico tools predict a damaging effect of the variant on protein function. The variant allele was found at a frequency of 2.6e-05 in 1607056 control chromosomes, predominantly at a frequency of 3.3e-05 within the Non-Finnish European subpopulation in the gnomAD database. This frequency is somewhat lower than the estimated maximum expected for a pathogenic variant in TP53 causing Li-Fraumeni Syndrome (4e-05), allowing no clear conclusions about variant significance. In addition, this variant was also reported in 1/7325 European American women, who were older than age 70, and have never had cancer (in the FLOSSIES database). The variant, c.877G>T, has been reported in the literature in an individual affected with glioblastoma who was diagnosed with neurofibromatosis type 1, however the variant was also found in an unaffected parent (Chung_1991), in addition the variant was reported in patients affected with breast cancer and other tumor phenotypes (e.g. Mitchell_2013, Tung_2016, Dorling_2021), however no supportive evidence for causality was provided. Publications reported experimental evidence evaluating an impact on protein function, and demonstrated that the variant protein was functional based on transcriptional activity in yeast (Kato_2003, Monti_2011), in addition, a loss-of-function saturation mutagenesis screen also indicated that this missense does not substantially affect TP53 function (Giacomelli_2018). The following publications have been ascertained in the context of this evaluation (PMID: 1686725, 3894400, 26976419, 33471991, 12826609, 30224644, 21343334). ClinVar contains an entry for this variant (Variation ID: 127826). Based on the evidence outlined above, the variant was classified as likely benign.
This missense variant replaces glycine with tryptophan at codon 293 of the TP53 protein. Computational prediction is inconclusive regarding the impact of this variant on protein structure and function. Functional studies have shown the mutant protein to be functional in yeast transcription activation assays (PMID: 12826609, 17606709, 21343334) and in human cell growth suppression assays (PMID: 30224644, 30840781). This variant has been reported in a 17 year-old affected with glioblastoma and diagnosed with neurofibromatosis type 1 (PMID: 1686725), individuals affected with adult-onset sarcoma (PMID: 23894400), and individuals affected with breast cancer (PMID: 26976419, 33471991). This variant has been identified in 3/282876 chromosomes in the general population by the Genome Aggregation Database (gnomAD). The available evidence is insufficient to determine the role of this variant in disease conclusively. Therefore, this variant is classified as a Variant of Uncertain Significance.
The NM_000546.6:c.877G>T variant in TP53 is a missense variant predicted to cause substitution of glycine by tryptophan at amino acid 293 (p.Gly293Trp). This variant has been observed in 4 heterozygous unrelated females from the same data source with no personal history of cancer prior to age 60 years and no personal history of sarcoma at any age (BS2_Moderate; Internal lab contributors: Ambry, SCV000187101.5). This variant has an allele frequency of 0.00003305 (39/1180036 alleles) in the European (Non-Finnish) population in gnomAD v4.1.0 which is lower than the Clingen TP53 VCEP threshold (<0.00004) for PM2_Supporting, and therefore meets this criterion (PM2_Supporting). In vitro assays performed in yeast and/or human cell lines showed functional transactivation and retained growth suppression activity indicating that this variant does not impact protein function(BS3; PMIDs: 12826609, 29979965, 30224644). In summary, TP53 c.877G>T (p.Gly293Trp) meets criteria to be classified as likely benign for Li-Fraumeni syndrome. ACMG/AMP criteria applied, as specified by the TP53 Expert Panel: BS2_Moderate, PM2_Supporting, BS3. (Bayesian Points: -5; VCEP specifications version 2.2, 1/16/2025).
Clinical Statement
This variant has been reported in ClinVar as Uncertain significance (5 clinical laboratories) and as Likely benign (3 clinical laboratories) and as Likely Benign by ClinGen TP53 Variant Curation Expert Panel, ClinGen expert panel.
Expert Panel Reviews
Likely Benign
ClinGen TP53 Variant Curation Expert Panel, ClinGen
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
No evidence of other pathogenic variants at position 293 in gene TP53
Functional Summary
The TP53 G293W variant, located within the DNA-binding domain of the Tp53 protein, results in a failure to induce apoptosis in Tp53-null cells in culture. This functional evidence supports a loss of Tp53 protein function.
Database Previews
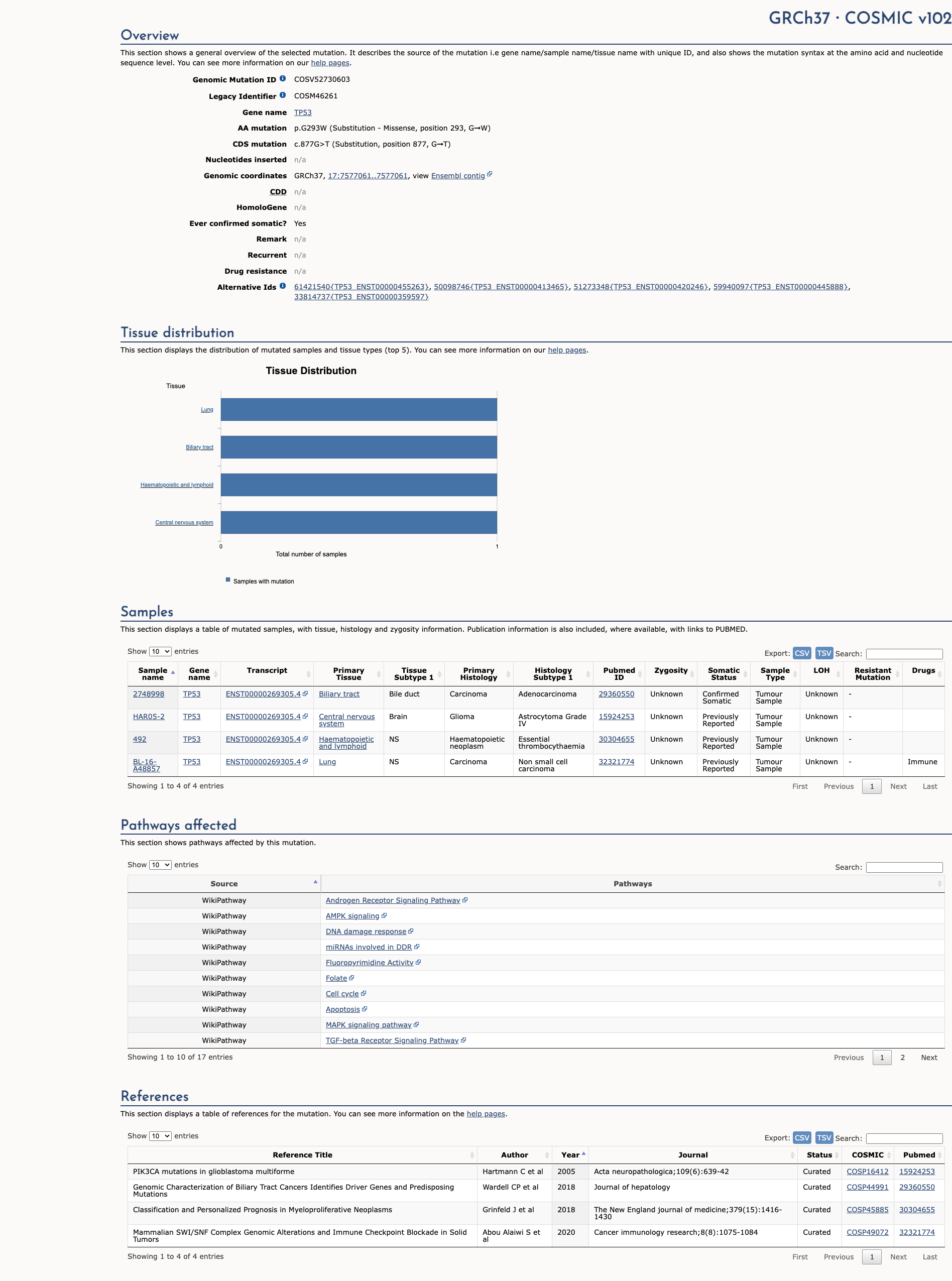
OncoKB
JAX-CKB
Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
REVEL Score
0.675
0.675
Likely Benign0.0
Uncertain (Low)0.2
Uncertain (Med)0.5
Likely Pathogenic0.75
REVEL scores ≥ 0.75 are strong evidence (PP3)
Predictor Consensus
Unknown
PP3 Applied
No
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Not Applied) Strength Modified
According to VCEP guidelines, PVS1 applies to null variants (nonsense, frameshift, canonical splice, deletions, duplications) predicted to undergo NMD or disrupt critical domains. The evidence for this variant shows it is a missense variant (p.G293W). Therefore, this criterion is not applied.
PS1
PS1 (Not Applied) Strength Modified
According to VCEP guidelines, PS1 can be applied when a variant results in the same amino acid change as a previously established pathogenic variant. The evidence for this variant shows no known pathogenic glycine-to-tryptophan change at codon 293 in TP53. Therefore, this criterion is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to VCEP guidelines, PS2 applies to confirmed de novo occurrences with points-based thresholds. There are no reported de novo observations for this variant. Therefore, this criterion is not applied.
PS3
PS3 (Not Applied) Strength Modified
According to VCEP guidelines, PS3 at any strength requires functional data from specific validated assays (Kato, Giacomelli, Kotler, Kawaguchi) demonstrating loss of function. The evidence for this variant shows failure to induce apoptosis in culture but lacks data from VCEP-specified assays. Therefore, this criterion is not applied.
PS4
PS4 (Not Applied) Strength Modified
According to VCEP guidelines, PS4 requires case counts converted to proband points for LFS-associated cancers. There are no case reports or proband points for this variant. Therefore, this criterion is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to VCEP guidelines, PM1 applies to missense variants at TP53 hotspot codons 175, 245, 248, 249, 273, or 282. The evidence for this variant shows it occurs at codon 293, outside these hotspots. Therefore, this criterion is not applied.
PM2
PM2 (Supporting) Strength Modified
According to VCEP guidelines, PM2_Supporting applies to variants with allele frequency <0.00003 in gnomAD. The evidence for this variant shows MAF = 0.00106% (0.0000106), which is below 0.00003. Therefore, this criterion is applied at Supporting strength.
PM3
PM3 (Not Applied) Strength Modified
According to standard ACMG, PM3 applies to detected variants in trans with a pathogenic variant for recessive disorders. This gene/disorder context and evidence are not applicable. Therefore, this criterion is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG, PM4 applies to protein length changes due to in-frame indels or stop-loss. This variant is a missense change without indel. Therefore, this criterion is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to VCEP guidelines, PM5 applies when other pathogenic missense variants affect the same residue. The evidence shows no other TP53 pathogenic variants reported at glycine 293. Therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to VCEP guidelines, PM6 applies to presumed de novo occurrences with limited evidence. No de novo data are available. Therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to VCEP guidelines, PP1 requires segregation in ≥3 meioses for supporting strength. No family segregation data are available. Therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG, PP2 applies to missense variants in genes with low rate of benign missense variation. TP53 is not characterized by low benign missense variation, and no VCEP recommendation was made. Therefore, this criterion is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to VCEP guidelines, PP3 at Moderate requires BayesDel ≥0.16 and no predicted splice impact; at Supporting requires BayesDel ≥0.16 and SpliceAI <0.2. BayesDel score is not available. Therefore, this criterion is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to VCEP guidelines, PP4 applies to observations of variant VAF in tumor studies. No such data exist. Therefore, this criterion is not applied.
PP5
PP5 (Not Applied) Strength Modified
According to standard ACMG, PP5 applies to reputable source assertions without evidence. ClinVar reports conflicting VUS and likely benign classifications. Therefore, this criterion is not applied.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, BA1 applies when allele frequency ≥0.001 in any gnomAD continental population. The evidence shows MAF = 0.00106% (<0.001). Therefore, this criterion is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines, BS1 applies when filtering allele frequency ≥0.0003 but <0.001. The evidence shows MAF = 0.0000106, which is below 0.0003. Therefore, this criterion is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to VCEP guidelines, BS2 applies to multiple older cancer-free females. No such data are available. Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines, BS3 requires functional assays showing retained function on Kato and another assay. The evidence shows loss of function. Therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to VCEP guidelines, BS4 applies to lack of segregation in affected family members. No segregation evidence is available. Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG, BP1 applies when missense changes occur in genes where only truncating variants cause disease. TP53 pathogenicity includes missense changes. Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG, BP2 applies to observed in cis or in trans with pathogenic variant in dominant gene. Not applicable in this context. Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG, BP3 applies to in-frame repetitive region indels. Not relevant for this missense variant. Therefore, this criterion is not applied.
BP4
BP4 (Not Applied) Strength Modified
According to VCEP guidelines, BP4 requires BayesDel <0.16 and SpliceAI <0.2. BayesDel score is not available. Therefore, this criterion is not applied.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG, BP5 applies when variant found in affected individual with alternative molecular basis for disease. No such context exists. Therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG, BP6 applies to unverified assertions by non-experts. ClinVar entries are VUS/likely benign by expert panel. Therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to VCEP guidelines, BP7 applies to silent or intronic variants outside splice sites. This is a missense variant. Therefore, this criterion is not applied.