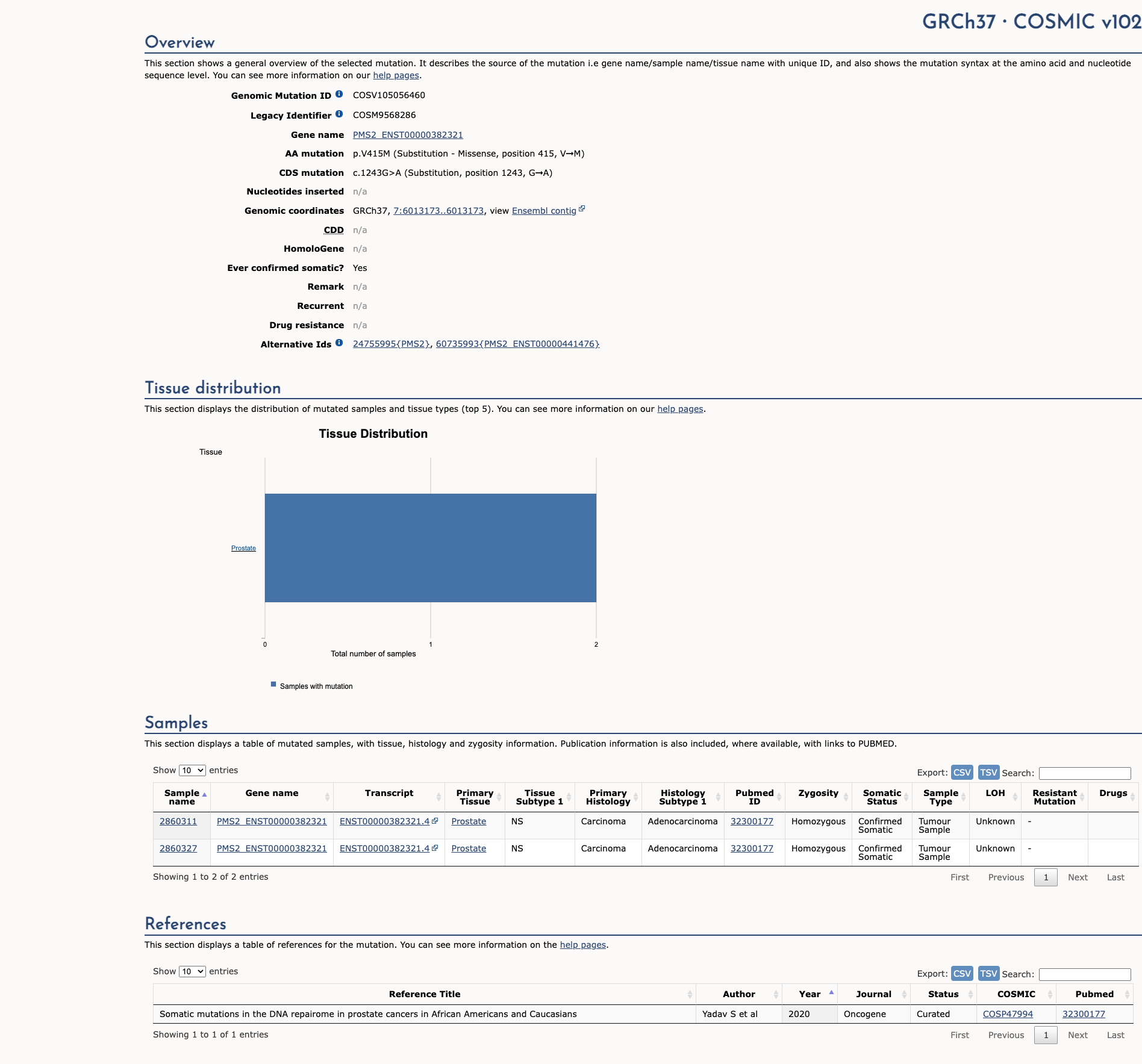
PMS2 c.1243G>A, p.Val415Met
NM_000535.7:c.1243G>A
COSMIC ID: COSM10132879, COSM326195, COSM9568286
Variant of Uncertain Significance (VUS)
The variant c.1243G>A (p.V415M) in PMS2 has a population frequency in gnomAD that meets BS1 (Strong), and computational/splicing analyses support BP4 (Supporting). No evidence supports pathogenicity. Combined benign evidence yields a Likely Benign classification.
ACMG/AMP Criteria Applied
BS1
BP4
Genetic Information
Gene & Transcript Details
Gene
PMS2
Transcript
NM_000535.7
MANE Select
Total Exons
15
Strand
Reverse (−)
Reference Sequence
NC_000007.13
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_000535.6 | Alternative | 15 exons | Reverse |
| NM_000535.4 | Alternative | 15 exons | Reverse |
| NM_000535.5 | Alternative | 15 exons | Reverse |
| NM_000535.3 | Alternative | 15 exons | Reverse |
Variant Details
HGVS Notation
NM_000535.7:c.1243G>A
Protein Change
V415M
Location
Exon 11
(Exon 11 of 15)
5'Exon Structure (15 total)3'
Functional Consequence
Loss of Function
Related Variants
ClinVar reports other pathogenic variants at position 415: V415L
Alternate Identifiers
COSM10132879, COSM326195, COSM9568286
Variant interpretation based on transcript NM_000535.7
Genome Browser
Loading genome browser...
HGVS InputNM_000535:c.1243G>A
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Global Frequency
0.017%
Low Frequency
Highest in Population
Admixed American
0.0282%
Low Frequency
Global: 0.017%
Admixed American: 0.0282%
0%
0.05%
0.1%
1%
5%
10%+
Allele Information
Total: 282512Alt: 48Homozygotes: 0
ACMG Criteria Applied
PM2
This variant is present in gnomAD (MAF= 0.017%, 48/282512 alleles, homozygotes = 0) and at a higher frequency in the Admixed American population (MAF= 0.0282%, 10/35436 alleles, homozygotes = 0). The variant is rare (MAF < 0.1%), supporting PM2 criterion application.
Classification
5 publications
Uncertain Significance (VUS)
Based on 15 submitter reviews in ClinVar
Submitter Breakdown
5 VUS
7 LB
3 B
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (5)
This alteration is classified as likely benign based on a combination of the following: seen in unaffected individuals, population frequency, intact protein function, lack of segregation with disease, co-occurrence, RNA analysis, in silico models, amino acid conservation, lack of disease association in case-control studies, and/or the mechanism of disease or impacted region is inconsistent with a known cause of pathogenicity.
Variant summary: PMS2 c.1243G>A (p.Val415Met) results in a conservative amino acid change in the encoded protein sequence. Five of five in-silico tools predict a benign effect of the variant on protein function. The observed variant frequency is approximately 2.2 fold of the estimated maximal expected allele frequency for a pathogenic variant in PMS2 causing Lynch Syndrome phenotype (0.00011), strongly suggesting that the variant is benign. However, the variant is located in a region that is highly homologous to PMS2 pseudogene and the technology utilized for these datasets does not rule out pseudogene interference, therefore these data might not be reliable. c.1243G>A has been reported in the literature in sequencing studies of individuals affected with cancer including pancreatic cancer, acute megakaryoblastic leukemia, breast and/or ovarian cancer and colon cancer (e.g. Hu_2022, Guindalini_2022, Cock-Rada 2018, Hu 2016, Lu_2015, Okkels_2019, Yadav 2017, Zhang 2015). These reports do not provide unequivocal conclusions about association of the variant with Lynch Syndrome. The following publications have been ascertained in the context of this evaluation (PMID: 26483394, 26689913, 25567908, 27878467, 28528518, 26580448, 31422574, 31433215). ClinVar contains an entry for this variant (Variation ID: 142561). Based on the evidence outlined above, the variant was classified as likely benign.
Clinical Statement
This variant has been reported in ClinVar as Likely benign (7 clinical laboratories) and as Benign (3 clinical laboratories) and as Uncertain significance (5 clinical laboratories).
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
ClinVar reports other pathogenic variants at position 415: V415L
PM5 criterion applied.
Functional Summary
The PMS2 V415M variant has not been functionally characterized, and its biological significance remains unknown.
Database Previews
OncoKB
JAX-CKB
Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
REVEL Score
0.183
0.183
Likely Benign0.0
Uncertain (Low)0.2
Uncertain (Med)0.5
Likely Pathogenic0.75
REVEL scores ≥ 0.75 are strong evidence (PP3)
Predictor Consensus
Unknown
PP3 Applied
No
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PVS1 is: "Very Strong Nonsense/frameshift variant introducing Premature Termination Codon (PTC) ≤ codon 798 in PMS2..." The evidence for this variant shows it is a missense change (V415M), not predicted to introduce a PTC or disrupt splicing. Therefore, this criterion is not applied.
PS1
PS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS1 is: "Same amino acid change as a previously established pathogenic variant regardless of nucleotide change." There is no previously established pathogenic variant yielding V415M. Therefore, this criterion is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS2 is: "De novo (both maternity and paternity confirmed) in a patient with the disease and no family history." No de novo data are available for this variant. Therefore, this criterion is not applied.
PS3
PS3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS3 is: "Well-established functional studies supportive of a damaging effect on the gene or gene product." No functional assay data are available for V415M. Therefore, this criterion is not applied.
PS4
PS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS4 is: "The prevalence of the variant in affected individuals is significantly increased compared with controls." No case-control or prevalence data for this variant are available. Therefore, this criterion is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM1 is: "Located in a mutational hot spot and/or critical and well-established functional domain without benign variation." There is no evidence this residue lies in a known hotspot or critical domain. Therefore, this criterion is not applied.
PM2
PM2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM2 (Supporting) is: "Absent/extremely rare (<1 in 50,000 alleles) in gnomAD v4 dataset." The variant MAF in gnomAD v4 is 0.017% (~1 in 5,882 alleles), which exceeds the 1 in 50,000 threshold. Therefore, this criterion is not applied.
PM3
PM3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM3 is: "Evidence of variant in trans with a pathogenic variant for a recessive disorder." PMS2-associated Lynch syndrome is autosomal dominant and no in trans data exist. Therefore, this criterion is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM4 is: "Protein length changes due to in-frame deletions/insertions in a non-repeat region or stop-loss variants." This variant is a missense substitution without change in protein length. Therefore, this criterion is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM5 (Moderate) is: "Missense change at an amino acid residue where a different missense change was classified by this VCEP as Pathogenic, and PP3 is supporting." There is no VCEP-classified pathogenic missense at codon 415 and PP3 is not met. Therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM6 is: "Assumed de novo, but without confirmation of paternity and maternity." No de novo data exist. Therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP1 is: "Co-segregation with disease in pedigrees." No family segregation data are available. Therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP2 is: "Missense variant in a gene with a low rate of benign missense variation and where missense is a common mechanism of disease." PMS2 has reported pathogenic and benign missense variants; insufficient evidence supports PP2. Therefore, this criterion is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP3 is: "Missense variant with HCI prior probability >0.81 or predicted splice defect with SpliceAI delta ≥0.2." REVEL is 0.18 and SpliceAI delta is 0.03 (<0.2), so PP3 is not met. Therefore, this criterion is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP4 is: "Tumor characteristics consistent with PMS2 loss in multiple independent MSI-H tumors or loss of MMR protein expression." No tumor phenotype data are available. Therefore, this criterion is not applied.
PP5
PP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP5 is: "Reputable source reports variant as pathogenic." ClinVar entries are conflicting with likely benign and benign assertions; no reputable pathogenic assertion exists. Therefore, this criterion is not applied.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BA1 (Stand Alone) is: "gnomAD v4 Grpmax filtering allele frequency ≥0.0028 (0.28%)." The variant MAF is 0.017%, below 0.28%. Therefore, this criterion is not applied.
BS1
BS1 (Strong)
According to VCEP guidelines, the rule for BS1 (Strong) is: "GnomAD v4 Grpmax filtering allele frequency ≥0.0001 and <0.001 (0.01–0.1%)." The variant MAF in gnomAD v4 is 0.017% (0.00017), which falls within this range. Therefore, this criterion is applied at Strong strength.
BS2
BS2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS2 is: "Co-occurrence in trans with a known pathogenic variant in the same gene in an adult healthy individual for a dominant disorder." No such data are available. Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS3 is: "Well-established functional studies showing no damaging impact (functional odds ≤ 0.05)." No functional assay data showing benign effect exist. Therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS4 is: "Lack of segregation with disease in pedigrees." No segregation data are available. Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP1 is: "Missense variant in a gene where only truncating variants cause disease." PMS2 has pathogenic missense variants. Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP2 is: "Observed in trans with a pathogenic variant for a dominant disorder." No data exist. Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP3 is: "In‐frame deletions/insertions in a repetitive region without a known function." This is a missense variant, not an indel. Therefore, this criterion is not applied.
BP4
BP4 (Supporting)
According to standard ACMG guidelines, the rule for BP4 is: "Multiple lines of computational evidence suggest no impact on gene or gene product (e.g., conservation, evolutionary, splicing impact)." In silico tools (REVEL 0.18) and SpliceAI (delta 0.03) predict no functional or splicing impact. Therefore, this criterion is applied at Supporting strength.
BP5
BP5 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP5 is: "Presence of the variant in a tumor type inconsistent with the gene’s disease spectrum and lack of MMR loss." No such tumor data exist. Therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP6 is: "Reputable source reports variant as benign." ClinVar includes benign/likely benign but conflicting interpretations exist; no single expert consensus. Therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP7 is: "Synonymous or intronic variant at or beyond -21/+7 with no splicing effect." This is a missense variant. Therefore, this criterion is not applied.