ATM c.6860G>A, p.Gly2287Glu
NM_000051.4:c.6860G>A
Variant of Uncertain Significance (VUS)
Only one benign supporting criterion (BP4) is met. No pathogenic criteria are met. Population frequency exceeds VCEP threshold for PM2 and PM5 cannot be applied. Insufficient evidence for classification beyond VUS.
ACMG/AMP Criteria Applied
BP4
Genetic Information
Gene & Transcript Details
Gene
ATM
Transcript
NM_000051.4
MANE Select
Total Exons
63
Strand
Forward (+)
Reference Sequence
NC_000011.9
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_000051.3 | RefSeq Select | 63 exons | Forward |
Variant Details
HGVS Notation
NM_000051.4:c.6860G>A
Protein Change
G2287E
Location
Exon 47
(Exon 47 of 63)
5'Exon Structure (63 total)3'
Functional Consequence
Loss of Function
Related Variants
ClinVar reports other pathogenic variants at position 2287: G2287A
Variant interpretation based on transcript NM_000051.4
Genome Browser
Loading genome browser...
HGVS InputNM_000051:c.6860G>A
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Global Frequency
0.00239%
Rare
Highest in Population
European (non-Finnish)
0.00528%
Rare
Global: 0.00239%
European (non-Finnish): 0.00528%
0%
0.05%
0.1%
1%
5%
10%+
Allele Information
Total: 251222Alt: 6Homozygotes: 0
ACMG Criteria Applied
PM2
This variant is present in gnomAD (MAF= 0.00239%, 6/251222 alleles, homozygotes = 0) and at a higher frequency in the European (non-Finnish) population (MAF= 0.00528%, 6/113564 alleles, homozygotes = 0). The variant is rare (MAF < 0.1%), supporting PM2 criterion application.
Classification
5 publications
Uncertain Significance (VUS)
Based on 14 submitter reviews in ClinVar
Submitter Breakdown
12 VUS
2 LB
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (5)
This variant was observed as part of a predisposition screen in an ostensibly healthy population. A literature search was performed for the gene, cDNA change, and amino acid change (where applicable). Publications were found based on this search. However, the evidence from the literature, in combination with allele frequency data from public databases where available, was not sufficient to rule this variant in or out of causing disease. Therefore, this variant is classified as a variant of unknown significance.
Variant summary: ATM c.6860G>A (p.Gly2287Glu) results in a non-conservative amino acid change located in the PIK-related kinase, FAT domain (IPR003151) of the encoded protein sequence. Three of five in-silico tools predict a benign effect of the variant on protein function. The variant allele was found at a frequency of 2.4e-05 in 251222 control chromosomes (genomAD). The available data on variant occurrences in the general population are insufficient to allow any conclusion about variant significance. c.6860G>A has been reported in the literature in individuals affected with kidney renal clear cell carcinoma and colorectal cancer (Lu_2015, Yurelun_2017). These reports do not provide unequivocal conclusions about association of the variant with Ataxia-Telangiectasia. To our knowledge, no experimental evidence demonstrating an impact on protein function has been reported. Seven ClinVar submitters (evaluation after 2014) cite the variant as uncertain significance. Based on the evidence outlined above, the variant was classified as uncertain significance.
This sequence change replaces glycine, which is neutral and non-polar, with glutamic acid, which is acidic and polar, at codon 2287 of the ATM protein (p.Gly2287Glu). This variant is present in population databases (rs1800061, gnomAD 0.006%). This missense change has been observed in individual(s) with colorectal cancer (PMID: 28135145). ClinVar contains an entry for this variant (Variation ID: 185012). Algorithms developed to predict the effect of missense changes on protein structure and function output the following: SIFT: "Not Available"; PolyPhen-2: "Benign"; Align-GVGD: "Not Available". The glutamic acid amino acid residue is found in multiple mammalian species, which suggests that this missense change does not adversely affect protein function. In summary, the available evidence is currently insufficient to determine the role of this variant in disease. Therefore, it has been classified as a Variant of Uncertain Significance.
This alteration is classified as likely benign based on a combination of the following: seen in unaffected individuals, population frequency, intact protein function, lack of segregation with disease, co-occurrence, RNA analysis, in silico models, amino acid conservation, lack of disease association in case-control studies, and/or the mechanism of disease or impacted region is inconsistent with a known cause of pathogenicity.
This missense variant replaces glycine with glutamic acid at codon 2287 of the ATM protein. Computational prediction suggests that this variant may not impact protein structure and function. To our knowledge, functional studies have not been reported for this variant. This variant has been reported in individuals affected with breast or colorectal cancer (PMID: 28135145, 28338653, 33471991). This variant has been identified in 6/251222 chromosomes in the general population by the Genome Aggregation Database (gnomAD). The available evidence is insufficient to determine the role of this variant in disease conclusively. Therefore, this variant is classified as a Variant of Uncertain Significance.
Clinical Statement
This variant has been reported in ClinVar as Uncertain significance (12 clinical laboratories) and as Likely benign (2 clinical laboratories).
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
ClinVar reports other pathogenic variants at position 2287: G2287A
PM5 criterion applied.
Functional Summary
The ATM G2287E variant has not been functionally characterized, and its biological significance remains unknown.
Database Previews
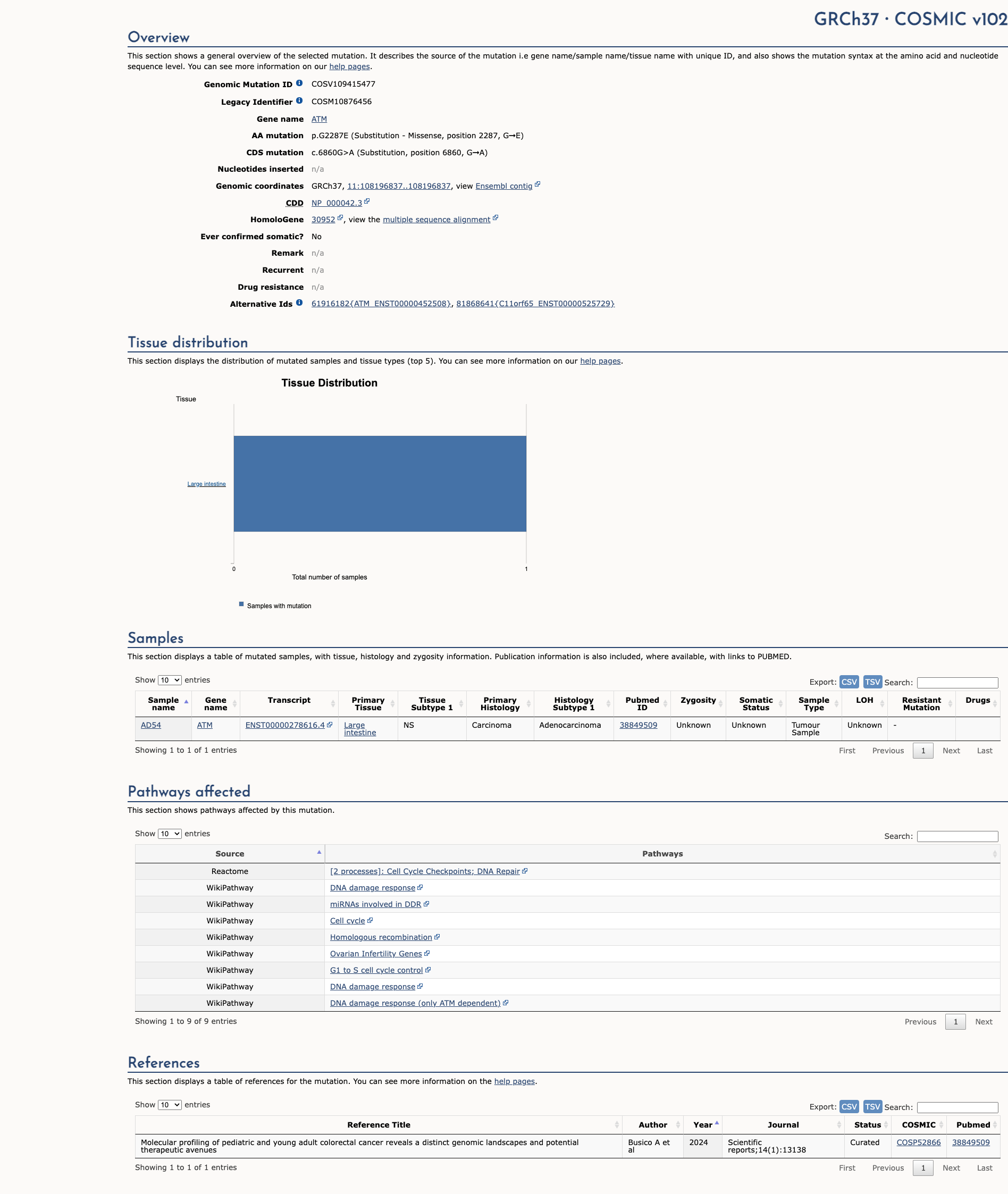
OncoKB
JAX-CKB
Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
REVEL Score
0.057
0.057
Likely Benign0.0
Uncertain (Low)0.2
Uncertain (Med)0.5
Likely Pathogenic0.75
REVEL scores ≥ 0.75 are strong evidence (PP3)
Predictor Consensus
Mixed/VUS
PP3 Applied
No
Additional Predictors
Benign:
CADD: 2.69polyphen_prediction: benignmetasvm: Tmetalr: Tprimateai: T
Neutral: Show all
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Not Applied) Strength Modified
According to VCEP guidelines (ClinGen-HBOP-ACMG-Specifications-ATM-version1_3.pdf), the rule for PVS1 is: 'Use ATM PVS1 Decision Tree'. The variant is a missense change and not a predicted null variant. Therefore, this criterion is not applied.
PS1
PS1 (Not Applied) Strength Modified
According to VCEP guidelines (ClinGen-HBOP-ACMG-Specifications-ATM-version1_3.pdf), the rule for PS1 is: 'Strong: Use for protein changes as long as splicing is ruled-out for both alterations.' There is no previously established pathogenic variant with the same amino acid change G2287E. Therefore, this criterion is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS2 is: 'De novo (both maternity and paternity confirmed) in a patient with the disease and no family history.' No de novo data are available for this variant. Therefore, this criterion is not applied.
PS3
PS3 (Not Applied) Strength Modified
According to VCEP guidelines (ClinGen-HBOP-ACMG-Specifications-ATM-version1_3.pdf), the rule for PS3 is gene-specific: functional studies must show failure to rescue ATM-specific features AND radiosensitivity. No functional assay data exist for G2287E. Therefore, this criterion is not applied.
PS4
PS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS4 is: 'Case-control studies; p-value ≤.05 AND (OR ≥2 or lower 95% CI ≥1.5).' No case-control or proband data are available. Therefore, this criterion is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM1 is: 'Located in a mutational hot spot and/or critical functional domain without benign variation.' G2287E is not reported in a known hotspot or critical domain. Therefore, this criterion is not applied.
PM2
PM2 (Not Applied) Strength Modified
According to VCEP guidelines (ClinGen-HBOP-ACMG-Specifications-ATM-version1_3.pdf), the rule for PM2_supporting is: 'Frequency ≤.001% if n=1 in a single sub population; n>1 in one or multiple subpopulations would not be considered rare.' The variant has MAF 0.00239% (6/251,222 alleles) with n>1. Therefore, this criterion is not applied.
PM3
PM3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM3 is recessive-specific, requiring detection in trans with pathogenic variant. No recessive inheritance data are available. Therefore, this criterion is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM4 is: 'Protein length changes due to in-frame indels or stop-loss.' G2287E is a missense change without length alteration. Therefore, this criterion is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to VCEP guidelines (ClinGen-HBOP-ACMG-Specifications-ATM-version1_3.pdf), the rule for PM5_supporting is: 'Use for genomic frameshift and truncating variants with PTC upstream of p.R3047.' G2287E is a missense variant. Therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM6 is: 'Assumed de novo without confirmation of paternity and maternity.' No parental testing data exist. Therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP1 is: 'Co-segregation with disease in multiple affected family members.' No segregation data are available. Therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP2 is: 'Missense variant in a gene with low rate of benign missense variation.' ATM has numerous benign missense variants. Therefore, this criterion is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to VCEP guidelines (ClinGen-HBOP-ACMG-Specifications-ATM-version1_3.pdf), the rule for PP3 is: 'Supporting if REVEL >0.7333 or RNA predictor shows splicing impact.' REVEL is 0.06 and SpliceAI is 0.01, indicating no impact. Therefore, this criterion is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP4 is: 'Patient’s phenotype or family history is highly specific for a disease with a single genetic etiology.' No phenotype data are provided. Therefore, this criterion is not applied.
PP5
PP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP5 (ClinVar assertions by reputable sources without evidence) is no longer recommended. Therefore, this criterion is not applied.
BA1
BA1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BA1 is: 'Allele frequency is >5%.' The variant’s MAF is 0.00239%, far below threshold. Therefore, this criterion is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS1 is: 'Strong: Filtering allele frequency >0.05%.' The variant’s MAF 0.00239% is below this. Therefore, this criterion is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS2 is: 'Observed in a healthy adult individual for a dominant disorder.' No such observations are reported. Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines (ClinGen-HBOP-ACMG-Specifications-ATM-version1_3.pdf), the rule for BS3 is: 'Moderate if rescues both ATM-specific feature and radiosensitivity; Supporting if rescues either.' No functional rescue data exist. Therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS4 is: 'Lack of segregation in affected members of a family.' No segregation data exist. Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP1 is: 'Missense variant in a gene where only truncating variants cause disease.' ATM pathogenic variants include missense changes. Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP2 is trans configuration evidence. No such data are available. Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP3 is: 'In-frame indel in repetitive region without known function.' G2287E is not an indel. Therefore, this criterion is not applied.
BP4
BP4 (Supporting)
According to VCEP guidelines (ClinGen-HBOP-ACMG-Specifications-ATM-version1_3.pdf), the rule for BP4 is: 'Supporting if REVEL ≤0.249 and RNA predictor shows no splicing impact.' This variant has REVEL 0.06 and SpliceAI 0.01. Therefore, this criterion is applied at Supporting strength.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP5 is: 'Variant found in a case with an alternate molecular basis for disease.' No such cases are reported. Therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP6 is: 'Reputable source reports variant as benign without evidence.' No low-confidence benign assertions are used. Therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP7 is: 'Synonymous variant with no predicted splice impact.' G2287E is missense. Therefore, this criterion is not applied.