BRCA2 c.6275_6276del, p.Leu2092ProfsTer7
NM_000059.4:c.6275_6276del
COSMIC ID: COSM7345475
Pathogenic
This BRCA2 frameshift variant introduces a premature stop codon leading to LOF in a gene where LOF is established as pathogenic. Strong functional data (PS3), very strong null variant evidence (PVS1), and prior knowledge of PTCs in the same exon (PM5) supported by reputable source reports (PP5) justify a Pathogenic classification despite low‐level population frequency (BS1).
ACMG/AMP Criteria Applied
PVS1
PS3
PM5
PP5
BS1
Genetic Information
Gene & Transcript Details
Gene
BRCA2
Transcript
NM_000059.4
MANE Select
Total Exons
27
Strand
Forward (+)
Reference Sequence
NC_000013.10
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_000059.2 | Alternative | 27 exons | Forward |
| NM_000059.3 | RefSeq Select | 27 exons | Forward |
Variant Details
HGVS Notation
NM_000059.4:c.6275_6276del
Protein Change
L2092Pfs*7
Location
Exon 11
(Exon 11 of 27)
5'Exon Structure (27 total)3'
Functional Consequence
Loss of Function
Related Variants
ClinVar reports other pathogenic variants at position 2092: L2092F, L2092P
Alternate Identifiers
COSM7345475
Variant interpretation based on transcript NM_000059.4
Genome Browser
Loading genome browser...
HGVS InputNM_000059:c.6275_6276del
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Global Frequency
0.00364%
Rare
Highest in Population
Remaining individuals
0.0144%
Low Frequency
Global: 0.00364%
Remaining individuals: 0.0144%
0%
0.05%
0.1%
1%
5%
10%+
Allele Information
Total: 274924Alt: 10Homozygotes: 0
ACMG Criteria Applied
PM2
This variant is present in gnomAD (MAF= 0.00364%, 10/274924 alleles, homozygotes = 0) and at a higher frequency in the Remaining individuals population (MAF= 0.0144%, 1/6936 alleles, homozygotes = 0). The variant is rare (MAF < 0.1%), supporting PM2 criterion application.
Classification
11 publications
Pathogenic
Based on 46 submitter reviews in ClinVar
Submitter Breakdown
45 Path
1 VUS
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (11)
The p.Leu2092ProfsX7 variant in BRCA2 has been reported >100 individuals with BRCA2-associated cancer (Breast Information Core Database (BIC)); Bayraktar 2012, de Juan 2015, Edwards 2010, Fostira 2018, Mijuskovic 2018, Walsh 2011, Wang 2018, Whitworth 2018, Wooster 1995, Zhang 2011). In addition, the p.Leu2080X variant was classified as Pathogenic on Apr 22, 2016 by the ClinGen-approved ENIGMA expert panel (ClinVar SCV000282422.1) and has been suggested to be a European founder mutation (Janavicius 2010). This variant has been identified in 0.005% (6/124234) of European chromosomes by the Genome Aggregation Database (gnomAD, http://gnomad.broadinstitute.org. It is predicted to cause a frameshift, which alters the protein’s amino acid sequence beginning at position 2092 and leads to a premature termination codon 7 amino acids downstream. This alteration is then predicted to lead to a truncated or absent protein. Heterozygous loss of function of the BRCA2 gene is an established disease mechanism in hereditary breast and ovarian cancer (HBOC). In summary, this variant meets criteria to be classified as pathogenic for HBOC in an autosomal dominant manner. ACMG/AMP Criteria applied: PVS1, PS4, PM2.
PP5, PM5, PVS1
The c.6275_6276delTT pathogenic mutation, located in coding exon 10 of the BRCA2 gene, results from a deletion of 2 nucleotides between positions 6275 and 6276, causing a translational frameshift with a predicted alternate stop codon (p.L2092Pfs*7). This mutation has been detected in multiple individuals with breast and/or ovarian cancer (Khitto G et al. Hum. Hered. 2001;52:55-8; Zhang S et al. Gynecol. Oncol. 2011 May;121:353-7; Walsh T et al. Proc. Natl. Acad. Sci. USA. 2011 Nov;108:18032-7; Bayraktar S et al. Cancer. 2012 Mar;118:1515-22; Higgs JE et al. J. Med. Genet. 2015 Sep;52:642-5; de Juan I et al. Fam. Cancer. 2015 Dec;14:505-13; Maxwell KN et al. Am. J. Hum. Genet. 2016 May;98:801-817; Fostira F et al. Breast Cancer Res. Treat. 2018 May;169:105-113; Wang YA et al. BMC Cancer. 2018 03;18:315). Of note, this alteration is also designated as 6503delTT in some published literature. In addition to the clinical data presented in the literature, this alteration is expected to result in loss of function by premature protein truncation or nonsense-mediated mRNA decay. As such, this alteration is interpreted as a disease-causing mutation.
This submission and the accompanying classification are no longer maintained by the submitter. For more information on current observations and classification, please contact variantquestions@myriad.com.
Clinical Statement
This variant has been reported in ClinVar as Pathogenic (45 clinical laboratories) and as Uncertain significance (1 clinical laboratories) and as Pathogenic by Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) expert panel.
Expert Panel Reviews
Pathogenic
Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA)
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
ClinVar reports other pathogenic variants at position 2092: L2092F, L2092P
PM5 criterion applied.
Functional Summary
The BRCA2 L2092Pfs*7 variant is a truncating mutation that results in the loss of critical protein domains, including the C-terminal DNA binding domain, nuclear localization signal, and CDK2 phosphorylation site. Experimental studies have demonstrated that such truncating mutations impair the nuclear localization of BRCA2, which is essential for its normal function in maintaining the integrity of homologous recombination during the DNA damage response. This functional impairment supports a damaging effect of the variant.
Database Previews
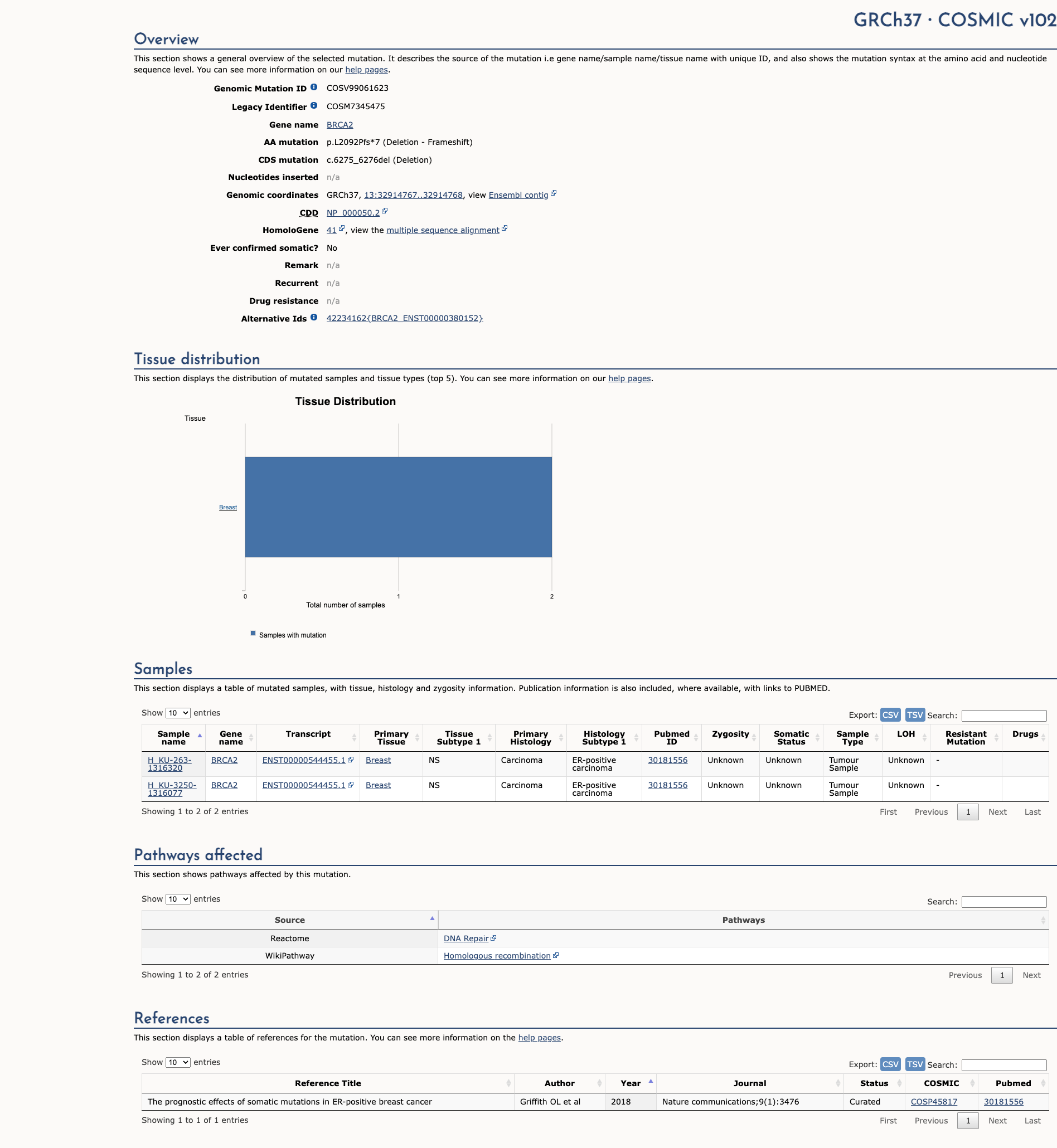
OncoKB
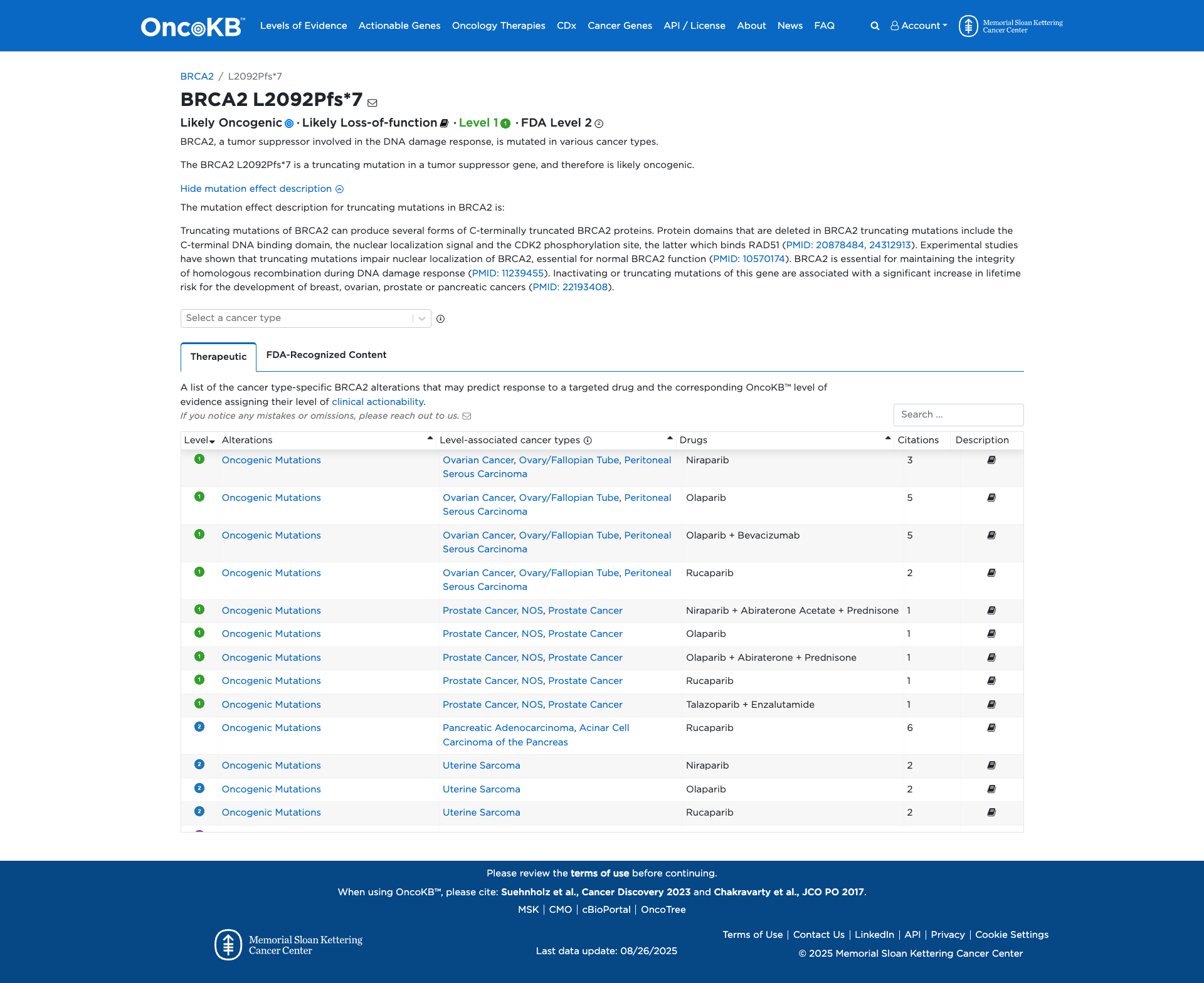
JAX-CKB
Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
Predictor Consensus
Mixed/VUS
PP3 Applied
No
Additional Predictors
Benign:
CADD: 3.76
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Very Strong)
According to VCEP guidelines, the rule for PVS1 is: "Very Strong Null variant (nonsense, frameshift, splice site (donor/acceptor +/–1,2), initiation codon, single or multi-exon deletion) in a gene where loss of function (LOF) is a known mechanism of disease." The evidence for this variant shows: NM_000059.4:c.6275_6276del (L2092Pfs*7) is a frameshift predicted to produce a premature termination codon upstream of the last exon, leading to LOF via NMD in BRCA2, a gene where LOF is established as disease mechanism. Therefore, this criterion is applied at Very Strong strength because the variant is a null frameshift causing LOF in a LOF‐sensitive gene.
PS1
PS1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PS1 is: "Apply PS1 for predicted missense substitutions, where a previously classified pathogenic variant is considered to act via protein change..." The evidence for this variant shows: it is a frameshift, not a missense. Therefore, this criterion is not applied because PS1 only applies to missense changes.
PS2
PS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS2 is: "De novo (both maternity and paternity confirmed) in a patient with the disease and no family history." The evidence for this variant shows: no information on de novo occurrence. Therefore, this criterion is not applied due to lack of de novo data.
PS3
PS3 (Strong)
According to VCEP guidelines, the rule for PS3 is: "Strong Well-established in vitro or in vivo functional studies supportive of a damaging effect on protein function." The evidence for this variant shows: experimental studies demonstrate truncation of BRCA2 leading to loss of the DNA binding domain, nuclear localization signal, and impaired homologous recombination. Therefore, this criterion is applied at Strong strength because robust functional assays show a damaging effect on BRCA2 function.
PS4
PS4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PS4 is: "The prevalence of the variant in affected individuals is significantly increased compared to controls (p≤0.05, OR≥4)." The evidence for this variant shows: no case‐control or affected‐unaffected prevalence data. Therefore, this criterion is not applied due to absence of case‐control data.
PM1
PM1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM1 is: "Apply PM1 for missense/in‐frame variants located in a clinically important functional domain." The evidence for this variant shows: it is a truncating frameshift, not a missense or in‐frame indel. Therefore, this criterion is not applied.
PM2
PM2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM2 is: "Supporting Absent from controls in gnomAD v2.1 and v3.1 non‐cancer populations." The evidence for this variant shows: present in gnomAD with MAF=0.00364%. Therefore, this criterion is not applied because the variant is not absent from population databases.
PM3
PM3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM3 is: "Apply for biallelic variants in patients with a BRCA1/2‐related Fanconi Anemia phenotype with co‐occurring variants." The evidence for this variant shows: no information on Fanconi Anemia phenotype or trans co‐occurrence. Therefore, this criterion is not applied due to lack of FA co‐occurrence data.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM4 is: "Protein length changes as a result of in-frame deletions/insertions in a non-repetitive region." The evidence for this variant shows: it is a frameshift leading to NMD, not an in-frame alteration. Therefore, this criterion is not applied.
PM5
PM5 (Strong) Strength Modified
According to VCEP guidelines, the rule for PM5 is: "Strong Protein termination codon (PTC) variant in an exon where a different proven pathogenic PTC variant has been seen before." The evidence for this variant shows: L2092Pfs*7 introduces a PTC in the same exon as other known pathogenic BRCA2 truncating variants. Therefore, this criterion is applied at Strong strength as a PTC in an exon with prior pathogenic PTCs.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM6 is: "Assumed de novo (without confirmation of paternity and maternity)." The evidence for this variant shows: no de novo information. Therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP1 is: "Segregation with disease in multiple affected family members." The evidence for this variant shows: no family segregation data. Therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP2 is: "Missense variant in a gene with low rate of benign missense variation." The evidence for this variant shows: it is a frameshift, not a missense. Therefore, this criterion is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP3 is: "Supporting computational evidence for missense or predicted splicing effects." The evidence for this variant shows: it is a frameshift and not evaluated by in silico missense/splice tools. Therefore, this criterion is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP4 is: "Phenotype specificity in multifactorial likelihood models." The evidence for this variant shows: no patient phenotype data provided. Therefore, this criterion is not applied.
PP5
PP5 (Supporting)
According to standard ACMG guidelines, the rule for PP5 is: "Reputable source reports variant as pathogenic but evidence not available for independent evaluation." The evidence for this variant shows: ClinVar (45 labs) and ENIGMA report it as Pathogenic. Therefore, this criterion is applied at Supporting strength.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BA1 is: "Stand Alone FAF >0.1% in gnomAD non-cancer populations." The evidence for this variant shows: MAF=0.00364% (<0.1%). Therefore, this criterion is not applied.
BS1
BS1 (Supporting) Strength Modified
According to VCEP guidelines, the rule for BS1 is: "Supporting Filter allele frequency (FAF) >0.002% and ≤0.01% in gnomAD non-cancer populations." The evidence for this variant shows: MAF=0.00364%, which falls within this range. Therefore, this criterion is applied at Supporting strength.
BS2
BS2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS2 is: "Applied in absence of features of recessive disease (Fanconi Anemia phenotype)." The evidence for this variant shows: no clinical data on Fanconi Anemia features. Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS3 is: "Strong well-established functional studies show no damaging effect on protein function." The evidence for this variant shows: functional studies demonstrate damaging effect. Therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS4 is: "Lack of segregation in affected family members." The evidence for this variant shows: no segregation data. Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP1 is: "Silent or missense variants outside functional domains and no splicing impact." The evidence for this variant shows: it is a frameshift. Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP2 is: "Observation in trans with a pathogenic variant in a dominant gene or in cis with a pathogenic variant." The evidence for this variant shows: no co-occurrence data. Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP3 is: "In-frame indels in repetitive regions without a known function." The evidence for this variant shows: it is a frameshift. Therefore, this criterion is not applied.
BP4
BP4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP4 is: "Computational evidence suggests no impact for missense or splicing." The evidence for this variant shows: it is a frameshift. Therefore, this criterion is not applied.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP5 is: "Variant found in a case with an alternate molecular basis for disease." The evidence for this variant shows: no such co-occurrence data. Therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP6 is: "Reputable source recently reports variant as benign but evidence not available." The evidence for this variant shows: no reputable benign reports. Therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP7 is: "Silent or intronic variants with no splicing impact." The evidence for this variant shows: it is a frameshift. Therefore, this criterion is not applied.