MSH2 c.2635C>T, p.Gln879Ter
NM_000251.2:c.2635C>T
Pathogenic
The c.2635C>T (p.Q879*) variant introduces a premature stop codon upstream of the VCEP-defined PTC threshold in MSH2, consistent with LOF. It is absent from population databases and reported by reputable sources as pathogenic, fulfilling PVS1 (Very Strong), PM2 (Supporting), and PP5 (Supporting), supporting a Pathogenic classification.
ACMG/AMP Criteria Applied
PVS1
PM2
PP5
Genetic Information
Gene & Transcript Details
Gene
MSH2
Transcript
NM_000251.3
MANE Select
Total Exons
16
Strand
Forward (+)
Reference Sequence
NC_000002.11
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_000251.2 | RefSeq Select | 16 exons | Forward |
| NM_000251.1 | Alternative | 16 exons | Forward |
Variant Details
HGVS Notation
NM_000251.2:c.2635C>T
Protein Change
Q879*
Location
Exon 16
(Exon 16 of 16)
5'Exon Structure (16 total)3'
Functional Consequence
Loss of Function
Related Variants
Variant interpretation based on transcript NM_000251.3
Genome Browser
Loading genome browser...
HGVS InputNM_000251:c.2635C>T
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Population Frequency
Global Frequency
0.0 in 100,000
Extremely Rare
Global: 0.0%
0%
0.05%
0.1%
1%
5%
10%+
ACMG Criteria Applied
PM2
This variant is not present in gnomAD (PM2 criteria applies).
Classification
1 publications
Pathogenic
Based on 4 submitter reviews in ClinVar
Submitter Breakdown
4 Path
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (1)
This sequence change creates a premature translational stop signal (p.Gln879*) in the MSH2 gene. RNA analysis indicates that this premature translational stop signal induces altered splicing and likely disrupts the C-terminus of the protein. This variant is not present in population databases (gnomAD no frequency). This premature translational stop signal has been observed in individual(s) with symptoms of Lynch syndrome (PMID: 15858146). This variant is also known as c.2637C>T. ClinVar contains an entry for this variant (Variation ID: 91028). Studies have shown that this premature translational stop signal results in skipping of exon 16 and introduces a new termination codon (Invitae). However the mRNA is not expected to undergo nonsense-mediated decay. This variant disrupts a region of the MSH2 protein in which other variant(s) (p.Leu888Cysfs*4) have been determined to be pathogenic (PMID: 8640829, 9222765). This suggests that this is a clinically significant region of the protein, and that variants that disrupt it are likely to be disease-causing. For these reasons, this variant has been classified as Pathogenic.
Clinical Statement
This variant has been reported in ClinVar as Pathogenic (4 clinical laboratories) and as Pathogenic by International Society for Gastrointestinal Hereditary Tumours (InSiGHT) expert panel.
Expert Panel Reviews
Pathogenic
International Society for Gastrointestinal Hereditary Tumours (InSiGHT)
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
Functional Summary
The MSH2 Q879* variant is a truncating mutation that likely results in loss of function of the MSH2 protein, a key component of the DNA mismatch repair pathway. Truncating mutations in MSH2 have been associated with Lynch syndrome and lead to impaired mismatch repair due to disruption of critical domains necessary for DNA binding and protein dimerization. This functional disruption supports a damaging effect of the variant.
Database Previews
OncoKB
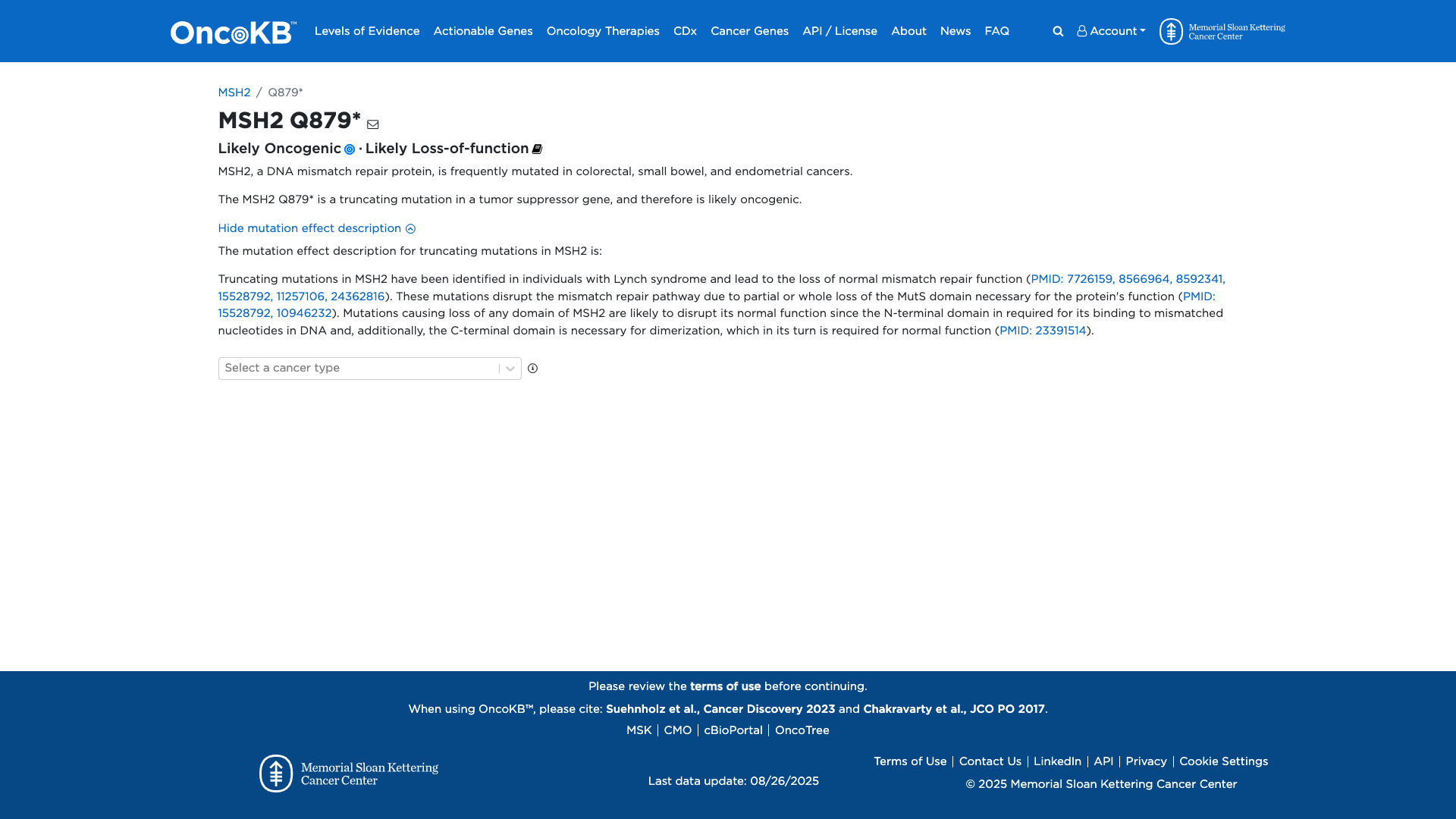
JAX-CKB
Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
Predictor Consensus
Mixed/VUS
PP3 Applied
No
Additional Predictors
Neutral: Show all
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Very Strong)
According to VCEP guidelines, "Very Strong Strength: Very Strong Nonsense/frameshift variant introducing Premature Termination Codon (PTC) ≤ codon 891 in MSH2." The evidence for this variant shows a nonsense change at codon 879 introducing a PTC within the threshold. Therefore, this criterion is applied at Very Strong strength because the variant meets the VCEP-defined threshold for a truncating LOF mutation.
PS1
PS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, PS1 applies when a missense substitution results in the same amino acid change as a known pathogenic variant. The evidence for this variant is a nonsense change (p.Q879*), not a missense change. Therefore, this criterion is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to VCEP guidelines, PS2 requires de novo occurrence with confirmation. No de novo testing or parental confirmation data are available. Therefore, this criterion is not applied.
PS3
PS3 (Not Applied) Strength Modified
According to VCEP guidelines, PS3 requires calibrated functional assays with quantified odds for pathogenicity or an MMR function defect confirmed by assays. No calibrated functional assay data are available for this variant. Therefore, this criterion is not applied.
PS4
PS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, PS4 requires case-control data demonstrating enrichment in affected individuals. No case-control studies are available for this variant. Therefore, this criterion is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM1 applies to variants located in a mutational hot spot or well‐studied functional domain without benign variation. There is no evidence that codon 879 lies in a defined hot spot or functional domain beyond LOF. Therefore, this criterion is not applied.
PM2
PM2 (Supporting) Strength Modified
According to VCEP guidelines, "Supporting Strength: Absent/extremely rare (<1 in 50,000 alleles) in gnomAD v4 dataset." The evidence shows the variant is absent from gnomAD (MAF=0%). Therefore, this criterion is applied at Supporting strength.
PM3
PM3 (Not Applied) Strength Modified
According to VCEP guidelines, PM3 applies to recessive disorders requiring evidence of allelic configuration. MSH2-related Lynch syndrome is autosomal dominant, and there is no evidence of trans configuration with another pathogenic variant. Therefore, this criterion is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM4 applies to in-frame indels or stop-loss variants altering protein length without LOF mechanism. This variant is a truncating nonsense change leading to LOF, not an in-frame indel. Therefore, this criterion is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM5 applies to novel missense changes at an amino acid residue where a different missense change is known to be pathogenic. This variant is a nonsense change, not missense. Therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM6 applies to assumed de novo variants without confirmation. No de novo or parental data are available. Therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP1 requires co-segregation data in families. No family segregation data are available. Therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP2 applies to missense variants in genes with low benign missense variation. This variant is a nonsense change, not missense. Therefore, this criterion is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP3 applies to multiple lines of computational evidence supporting a deleterious effect. In silico predictions are mixed and do not uniformly support deleterious impact. Therefore, this criterion is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to VCEP guidelines, PP4 requires tumor phenotype (MSI-H, loss of protein expression) consistent with gene defect. No tumor phenotype or IHC data are provided. Therefore, this criterion is not applied.
PP5
PP5 (Supporting)
According to standard ACMG guidelines, "Reputable source reports variant as pathogenic but evidence is not available for independent evaluation." ClinVar (4 clinical labs) and InSiGHT expert panel report this variant as Pathogenic. Therefore, this criterion is applied at Supporting strength.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, BA1 applies if MAF ≥0.1% in gnomAD v4. The variant MAF=0% in gnomAD. Therefore, this criterion is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines, BS1 applies if MAF ≥0.01% and <0.1%. The variant MAF=0%, which is below threshold. Therefore, this criterion is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to VCEP guidelines, BS2 requires co-occurrence in trans with a known pathogenic variant and clinical assessment. No such data are available. Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to standard ACMG guidelines, BS3 requires well-established functional assays showing no damaging effect. No such data are available. Therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, BS4 requires lack of segregation in families. No segregation data are available. Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP1 applies to missense variants in genes where only LOF causes disease. This is a truncating LOF variant, not missense. Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP2 requires observation in trans with a pathogenic variant in recessive disorders. MSH2 is autosomal dominant, and no such data exist. Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP3 applies to in-frame indels in repetitive regions. The variant is a nonsense change, not an in-frame indel. Therefore, this criterion is not applied.
BP4
BP4 (Not Applied) Strength Modified
According to VCEP guidelines, BP4 applies to in silico predictions showing no impact for intronic or synonymous variants. The variant is a nonsense change, and computational evidence is not uniformly benign. Therefore, this criterion is not applied.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP5 applies when a variant is found in a case with an alternative molecular cause. No such evidence is available. Therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP6 applies when a reputable source reports the variant as benign. No such reports exist. Therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP7 applies to synonymous or intronic variants without splicing impact. The variant is a nonsense change, not synonymous/intronic. Therefore, this criterion is not applied.

