TP53 c.742C>G, p.Arg248Gly
NM_000546.5:c.742C>G
COSMIC ID: COSM1189383, COSM2744594
Pathogenic
The R248G variant in TP53 meets PS3_Strong, PM1_Moderate, PM5_Moderate, PM2_Supporting, and PP5_Supporting, consistent with Pathogenic classification under TP53 VCEP specifications.
ACMG/AMP Criteria Applied
PS3
PM1
PM2
PM5
PP5
Genetic Information
Gene & Transcript Details
Gene
TP53
Transcript
NM_000546.6
MANE Select
Total Exons
11
Strand
Reverse (−)
Reference Sequence
NC_000017.10
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_000546.5 | RefSeq Select | 11 exons | Reverse |
| NM_000546.3 | Alternative | 11 exons | Reverse |
| NM_000546.4 | Alternative | 11 exons | Reverse |
| NM_000546.2 | Alternative | 11 exons | Reverse |
Variant Details
HGVS Notation
NM_000546.5:c.742C>G
Protein Change
R248G
Location
Exon 7
(Exon 7 of 11)
5'Exon Structure (11 total)3'
Functional Consequence
Loss of Function
Related Variants
ClinVar reports other pathogenic variants at position 248: R248Q, R248W
Alternate Identifiers
COSM1189383, COSM2744594
Variant interpretation based on transcript NM_000546.6
Genome Browser
Loading genome browser...
HGVS InputNM_000546:c.742C>G
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Population Frequency
Global Frequency
0.0 in 100,000
Extremely Rare
Global: 0.0%
0%
0.05%
0.1%
1%
5%
10%+
ACMG Criteria Applied
PM2
This variant is not present in gnomAD (PM2 criteria applies).
Classification
3 publications
Likely Pathogenic
Based on 4 submitter reviews in ClinVar
Submitter Breakdown
1 Path
3 LP
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (3)
This variant is considered likely pathogenic. Functional studies indicate this variant impacts protein function [PMID: 29979965]. This variant is expected to disrupt protein structure [Myriad internal data].
The p.R248G pathogenic mutation (also known as c.742C>G), located in coding exon 6 of the TP53 gene, results from a C to G substitution at nucleotide position 742. The arginine at codon 248 is replaced by glycine, an amino acid with dissimilar properties. This variant is in the DNA binding domain of the TP53 protein and was shown to have loss of transactivation capacity in yeast-based functional studies (Kato S et al. Proc Natl Acad Sci USA. 2003 Jul 8;100(14):8424-9). In addition, studies conducted in human cell lines indicate this alteration is deficient at growth suppression and has a dominant negative effect (Kotler E et al. Mol.Cell. 2018 Jul;71:178-190.e8; Giacomelli AO et al. Nat. Genet. 2018 Oct;50:1381-1387). This specific alteration has not been reported as a germline finding in the literature, although several other alterations at this same codon (p.R248Q, p.R248W and p.R248L) have been reported in Li-Fraumeni Syndrome (LFS), or LFS-related cancers (Petitjean A et al. IARC TP53 database [version R15, November 2010]. Hum Mutat. 2007 Jun;28(6):622-9; Rausch T et al. Cell. 2012 Jan; 148(1-2):59-71; Toguchida J et al. N. Engl. J. Med. 1992 May; 326(20):1301-8; Rieber J et al. Genes Chromosomes Cancer. 2009 Jul; 48(7):558-68). This amino acid position is highly conserved in available vertebrate species. In addition, this alteration is predicted to be deleterious by in silico analysis. Based on the supporting evidence, this alteration is interpreted as a disease-causing mutation.
This sequence change replaces arginine, which is basic and polar, with glycine, which is neutral and non-polar, at codon 248 of the TP53 protein (p.Arg248Gly). This variant is not present in population databases (gnomAD no frequency). This missense change has been observed in individual(s) with clinical features of Li-Fraumeni syndrome (Invitae). ClinVar contains an entry for this variant (Variation ID: 376652). Advanced modeling performed at Invitae incorporating data from internal and/or published experimental studies (PMID: 12826609, 29979965, 30224644) indicates that this missense variant is expected to disrupt TP53 function with a positive predictive value of 97.5%. Experimental studies have shown that this missense change affects TP53 function (PMID: 9290701, 11920959, 12826609, 22919068, 29979965, 30224644). In summary, the currently available evidence indicates that the variant is pathogenic, but additional data are needed to prove that conclusively. Therefore, this variant has been classified as Likely Pathogenic.
Clinical Statement
This variant has been reported in ClinVar as Likely pathogenic (3 clinical laboratories) and as Pathogenic (1 clinical laboratories).
COSMIC ID
COSM1189383, COSM2744594
Recurrence
37 occurrences
PM1 Criteria
Applied
Criterion PM1 is applied based on the high recurrence in COSMIC database.
COSMIC Database Preview
Accessing full COSMIC database details requires institutional login or subscription. External links may prompt for authentication.
Functional Impact
Functional Domain
Hotspot Status
Hotspot
PM1
Mutation Count
3360
Reported mutations in this domain
050100+
Domain Summary
This variant is located in a mutational hotspot or critical domain (3360 mutations).
PM1 criterion applied.
Related Variants in This Domain
ClinVar reports other pathogenic variants at position 248: R248Q, R248W
PM5 criterion applied.
Functional Summary
The TP53 R248G variant has been functionally characterized and is associated with a loss of function. In vivo studies in yeast and in vitro studies in human cancer cell lines have demonstrated that this mutation results in the loss of transactivational and growth suppression activities compared to the wildtype. Additionally, while the variant retains some ability to transactivate certain target genes, it fails to repress specific transcriptional activities, further supporting its role in loss of TP53 protein function.
Database Previews
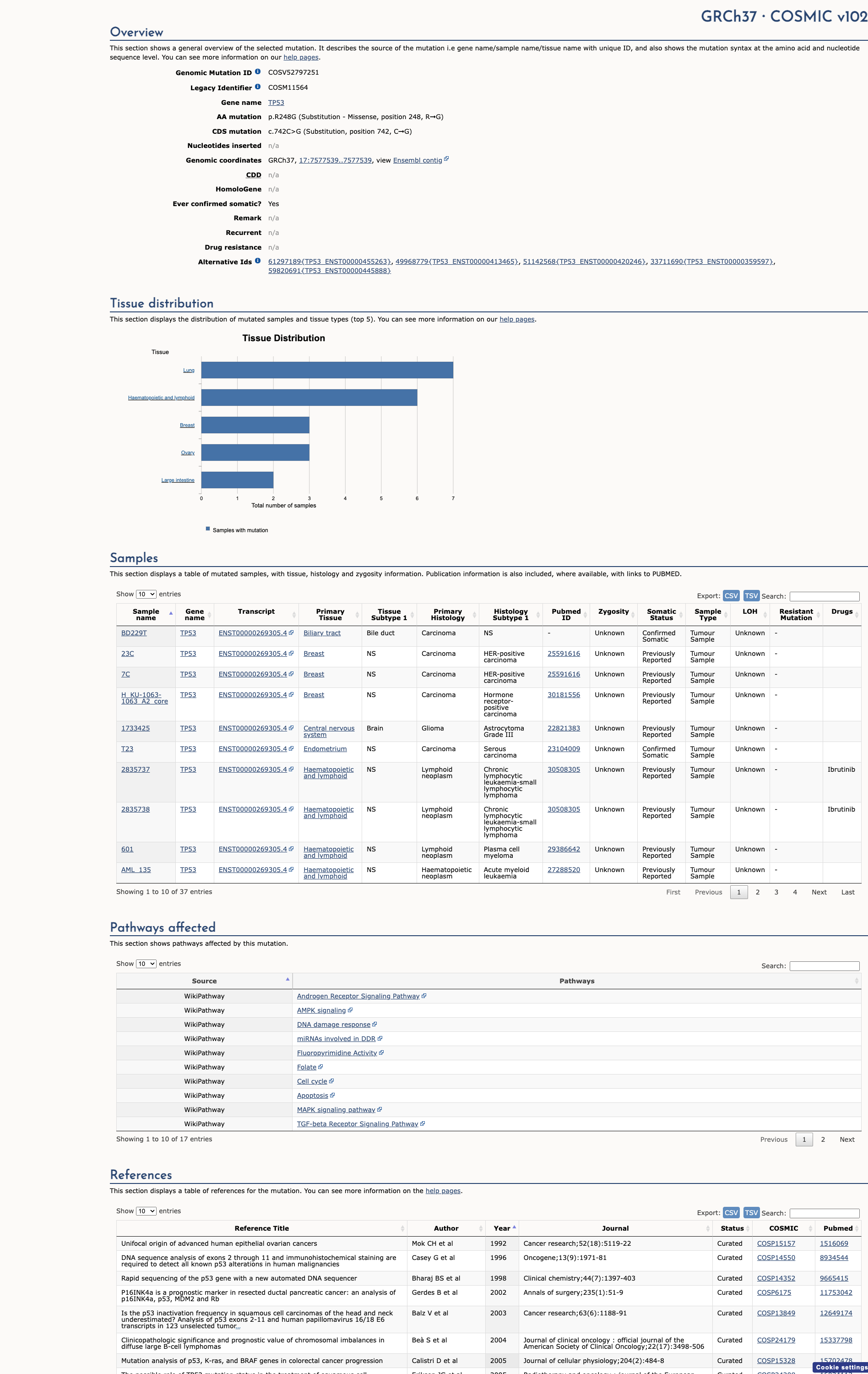
OncoKB
JAX-CKB
Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
REVEL Score
0.932
0.932
Likely Benign0.0
Uncertain (Low)0.2
Uncertain (Med)0.5
Likely Pathogenic0.75
REVEL scores ≥ 0.75 are strong evidence (PP3)
Predictor Consensus
Mixed/VUS
PP3 Applied
Yes
Additional Predictors
Pathogenic:
metasvm: Dmetalr: D
Benign:
CADD: 5.28primateai: T
Neutral: Show all
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Not Applied) Strength Modified
According to VCEP guidelines, PVS1 applies to null variants (nonsense, frameshift, canonical ±1,2 splice site, initiation codon) predicted to result in NMD. The evidence for this variant shows that NM_000546.5:c.742C>G (R248G) is a missense change, not a null variant. Therefore, PVS1 is not applied.
PS1
PS1 (Not Applied) Strength Modified
According to VCEP guidelines, PS1 applies when the same amino acid change as a known pathogenic variant occurs via a different nucleotide change. The evidence for this variant shows no prior report of R248G via a different nucleotide change. Therefore, PS1 is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to VCEP guidelines, PS2 requires confirmed de novo occurrence with parental testing. No de novo data are available for this variant. Therefore, PS2 is not applied.
PS3
PS3 (Strong)
According to VCEP guidelines, PS3_Strong: "Non-functional on Kato et al. data AND loss of function (LOF) on another assay". The evidence for R248G shows loss of transactivation and growth suppression in yeast (Kato et al.) and human cell line assays. Therefore, PS3 is applied at Strong strength.
PS4
PS4 (Not Applied) Strength Modified
According to VCEP guidelines, PS4 requires quantitative proband or segregation data demonstrating significant enrichment in affected individuals. No such data are provided. Therefore, PS4 is not applied.
PM1
PM1 (Moderate)
According to VCEP guidelines, PM1_Moderate: "Missense variants within the following codons using transcript NM_00546.4: 175, 245, 248, 249, 273, 282." The evidence for R248G places it at codon 248, a known hotspot. Therefore, PM1 is applied at Moderate strength.
PM2
PM2 (Supporting) Strength Modified
According to VCEP guidelines, PM2_Supporting: "Variant allele frequency <0.00003 in gnomAD or other large population database." The evidence for R248G shows absence from gnomAD (MAF=0%). Therefore, PM2 is applied at Supporting strength.
PM3
PM3 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM3 applies to variants observed in trans with a pathogenic variant in a recessive disorder. TP53-associated disease is dominant and no trans data exist. Therefore, PM3 is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM4 applies to protein length–altering variants. R248G is a missense change with no length alteration. Therefore, PM4 is not applied.
PM5
PM5 (Moderate)
According to VCEP guidelines, PM5_Moderate: "Missense variant at an amino acid residue where 1 different missense variant previously determined to be pathogenic has been seen before." The evidence for R248G shows R248W is a known pathogenic missense at the same residue. Therefore, PM5 is applied at Moderate strength.
PM6
PM6 (Not Applied) Strength Modified
According to VCEP guidelines, PM6 applies to assumed de novo variants without confirmation. No de novo data are available. Therefore, PM6 is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to VCEP guidelines, PP1 requires cosegregation data. No segregation data are provided. Therefore, PP1 is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP2 applies to missense variants in genes with low rates of benign missense variation. TP53 has both benign and pathogenic missense variants and this criterion is not specific. Therefore, PP2 is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to VCEP guidelines, PP3 requires aGVGD Class C65 AND BayesDel score ≥0.16 for Moderate or aGVGD C25–C55 AND BayesDel ≥0.16 for Supporting. The evidence for R248G includes REVEL and MetaSVM/MetaLR but BayesDel and aGVGD data are unavailable. Therefore, PP3 is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to VCEP guidelines, PP4 applies to tumor VAF observations. No VAF data in a clinical context are provided. Therefore, PP4 is not applied.
PP5
PP5 (Supporting)
According to standard ACMG guidelines, PP5: "Reputable source recently reports variant as pathogenic but evidence is not available for independent evaluation." The evidence for R248G shows ClinVar reports by multiple laboratories as Likely Pathogenic/Pathogenic. Therefore, PP5 is applied at Supporting strength.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, BA1 applies to variants with filtering allele frequency ≥0.001. R248G is absent from population databases. Therefore, BA1 is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines, BS1 applies to variants with filtering allele frequency ≥0.0003. R248G is absent from population databases. Therefore, BS1 is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to VCEP guidelines, BS2 requires observation of ≥2 unrelated unaffected female carriers aged ≥60. No such data are provided. Therefore, BS2 is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines, BS3 applies to functional assays showing no loss of function. The evidence for R248G shows loss of function. Therefore, BS3 is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to VCEP guidelines, BS4 requires lack of segregation in affected members. No segregation data are provided. Therefore, BS4 is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP1 applies to missense variants in genes where only truncating variants cause disease. TP53 has many pathogenic missense variants. Therefore, BP1 is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP2 applies to variants observed in cis with a pathogenic variant in a dominant disorder. No such data are provided. Therefore, BP2 is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP3 applies to in-frame indels in repetitive regions. R248G is a missense variant. Therefore, BP3 is not applied.
BP4
BP4 (Not Applied) Strength Modified
According to VCEP guidelines, BP4 requires BayesDel <0.16 AND no predicted splice impact (SpliceAI <0.2). BayesDel data are unavailable. Therefore, BP4 is not applied.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP5 applies when a variant is found in a case with an alternate molecular basis for disease. No such data are provided. Therefore, BP5 is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP6 applies when a reputable source reports a variant as benign without evidence. No benign reports exist. Therefore, BP6 is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to VCEP guidelines, BP7 applies to synonymous or intronic variants outside splice regions. R248G is a missense variant. Therefore, BP7 is not applied.