PTEN c.493-1G>A, p.Splice_Site
NM_000314.8:c.493-1G>A
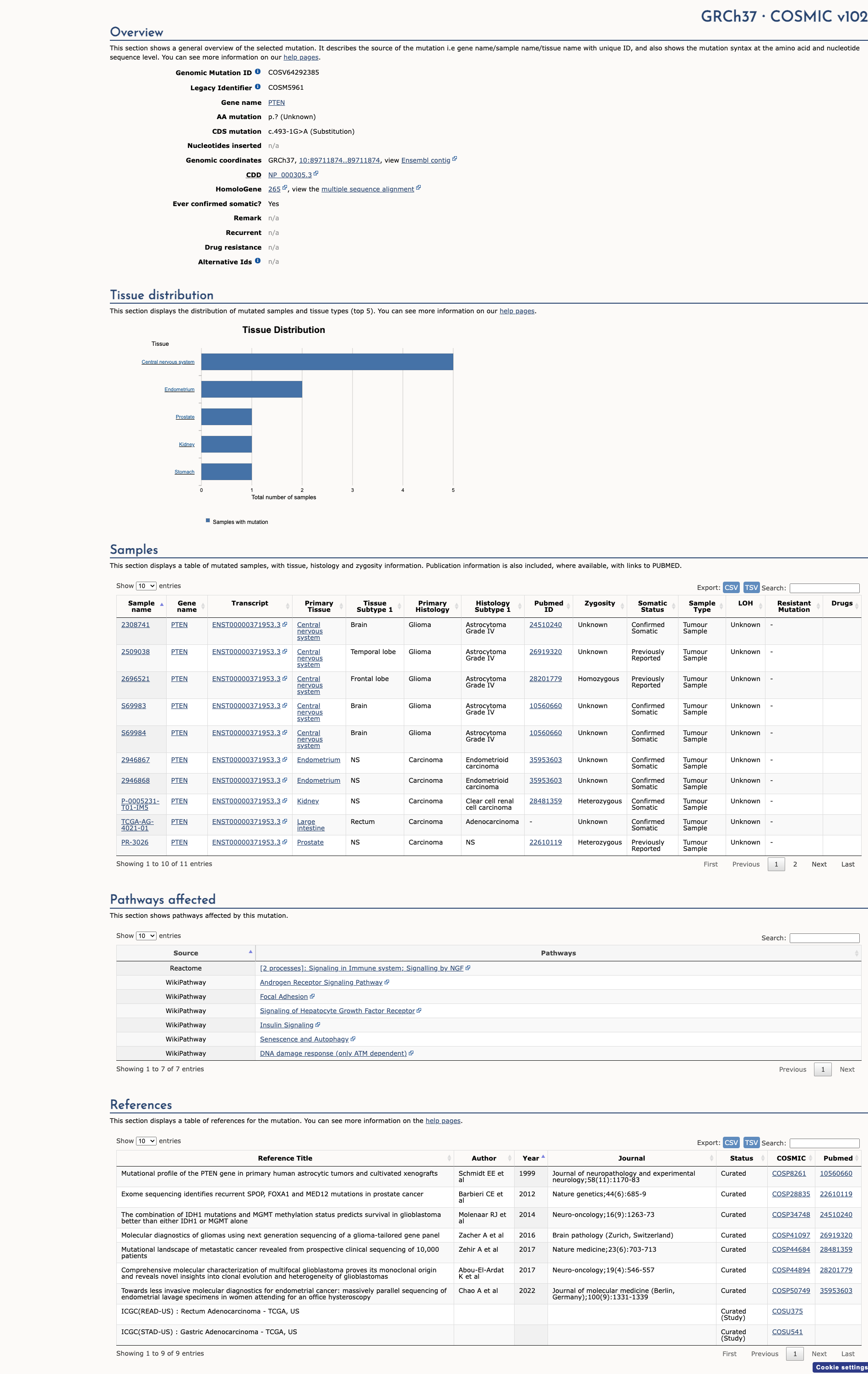
COSMIC ID: COSM5961
Pathogenic
The canonical splice acceptor variant c.493-1G>A in PTEN is absent from population databases (PM2), predicted to abolish normal splicing (PVS1, PP3), and reported as pathogenic in ClinVar (PP5). Applying VCEP PTEN rules yields PVS1 Very Strong, PM2 Supporting, PP3 Supporting, PP5 Supporting, consistent with a Pathogenic classification.
ACMG/AMP Criteria Applied
PVS1
PM2
PP3
PP5
Genetic Information
Gene & Transcript Details
Gene
PTEN
Transcript
NM_000314.8
MANE Select
Total Exons
9
Strand
Forward (+)
Reference Sequence
NC_000010.10
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_000314.7 | RefSeq Select | 9 exons | Forward |
| NM_000314.5 | Alternative | 9 exons | Forward |
| NM_000314.4 | Alternative | 9 exons | Forward |
| NM_000314.3 | Alternative | 9 exons | Forward |
| NM_000314.6 | Alternative | 9 exons | Forward |
Variant Details
HGVS Notation
NM_000314.8:c.493-1G>A
Protein Change
Splice
Location
Exon 5
(Exon 5 of 9)
5'Exon Structure (9 total)3'
Functional Consequence
Loss of Function
Related Variants
Alternate Identifiers
COSM5961
Variant interpretation based on transcript NM_000314.8
Genome Browser
Loading genome browser...
HGVS InputNM_000314:c.493-1G>A
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Population Frequency
Global Frequency
0.0 in 100,000
Extremely Rare
Global: 0.0%
0%
0.05%
0.1%
1%
5%
10%+
ACMG Criteria Applied
PM2
This variant is not present in gnomAD (PM2 criteria applies).
Classification
3 publications
Pathogenic
Based on 3 submitter reviews in ClinVar
Submitter Breakdown
3 Path
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (3)
The c.493-1G>A intronic pathogenic mutation results from a G to A substitution one nucleotide upstream from coding exon 6 of the PTEN gene. This alteration has been identified in individuals with PTEN-related disease (Ambry internal data; De Rubeis S et al. Nature, 2014 Nov;515:209-15; Wang T et al. Nat Commun, 2016 11;7:13316; Kosmicki JA et al. Nat. Genet., 2017 Apr;49:504-510). This variant is considered to be rare based on population cohorts in the Genome Aggregation Database (gnomAD). In silico splice site analysis predicts that this alteration will weaken the native splice acceptor site and will result in the creation or strengthening of a novel splice acceptor site; however, direct evidence is insufficient at this time (Ambry internal data). In addition to the clinical data, alterations that disrupt the canonical splice site are expected to cause aberrant splicing, resulting in an abnormal protein or a transcript that is subject to nonsense-mediated mRNA decay. As such, this alteration is classified as a disease-causing mutation.
Algorithms developed to predict the effect of sequence changes on RNA splicing suggest that this variant may disrupt the consensus splice site. For these reasons, this variant has been classified as Pathogenic. ClinVar contains an entry for this variant (Variation ID: 189409). Disruption of this splice site has been observed in individuals with PTEN hamartoma tumor syndrome (PMID: 17526801, 28677221). This variant is not present in population databases (gnomAD no frequency). This sequence change affects an acceptor splice site in intron 5 of the PTEN gene. It is expected to disrupt RNA splicing. Variants that disrupt the donor or acceptor splice site typically lead to a loss of protein function (PMID: 16199547), and loss-of-function variants in PTEN are known to be pathogenic (PMID: 9467011, 21194675).
Clinical Statement
This variant has been reported in ClinVar as Pathogenic (3 clinical laboratories).
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
Computational Analysis
Pathogenicity Predictions
Predictor Consensus
Mixed/VUS
PP3 Applied
No
Additional Predictors
Benign:
CADD: 8.25
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Very Strong)
According to VCEP guidelines, the rule for PVS1 is: 'Null Variant (nonsense, fs, start codon, splicing +1/2, startgain, single or multi-exon deletion) in a gene where LOF is a known mechanism of disease' at Very Strong strength. The evidence for this variant shows a canonical splice acceptor site mutation (c.493-1G>A) predicted to abolish normal splicing, not in the last exon. Therefore, this criterion is applied at Very Strong strength because a +/−1 canonical splice variant is predicted to result in loss of function in PTEN.
PS1
PS1 (Not Applied) Strength Modified
According to VCEP guidelines, PS1 requires a different variant at the same nucleotide as a known pathogenic splicing variant with equal or greater in silico impact. There is no previously established pathogenic variant at c.493-1. Therefore, PS1 is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to VCEP guidelines, PS2 requires de novo observation(s) with appropriate confirmation. No de novo data are provided for this variant. Therefore, PS2 is not applied.
PS3
PS3 (Not Applied) Strength Modified
According to PTEN pre-processing, the variant was not found in the functional assay dataset (Table S2-Table 1.csv); no well-established in vitro or in vivo functional studies are available. Therefore, PS3 is not applied.
PS4
PS4 (Not Applied) Strength Modified
According to VCEP guidelines, PS4 requires case-control data or proband specificity scores. No prevalence or proband specificity data are provided. Therefore, PS4 is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to VCEP guidelines, PM1 applies to variants in critical catalytic motifs (residues 90-94, 123-130, 166-168). This splice variant is outside those regions. Therefore, PM1 is not applied.
PM2
PM2 (Supporting) Strength Modified
According to VCEP guidelines, PM2 is: 'Absent in population databases present at <0.001% allele frequency in gnomAD'. The evidence shows the variant is absent from gnomAD. Therefore, PM2 is applied at Supporting strength because the allele is not observed in large population datasets.
PM3
PM3 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM3 applies to recessive inheritance with variants in trans. PTEN is associated with autosomal dominant disease, and no trans observations are reported. Therefore, PM3 is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM4 applies to in-frame indels or stop-loss variants. This is a splice-site variant, not an in-frame indel. Therefore, PM4 is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to VCEP guidelines, PM5 applies to missense changes at residues with known pathogenic missense variants. This is a splice acceptor variant, not missense. Therefore, PM5 is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to VCEP guidelines, PM6 requires assumed de novo occurrence without confirmation. No presumed de novo data are available. Therefore, PM6 is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to VCEP guidelines, PP1 requires co-segregation in multiple affected family members. No segregation data are provided. Therefore, PP1 is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP2 applies to missense variants in genes with low benign missense variation. This is a splice variant, not missense. Therefore, PP2 is not applied.
PP3
PP3 (Supporting)
According to VCEP guidelines, PP3 for splicing variants is: 'Concordance of SpliceAI and VarSeak predictions.' SpliceAI predicts high impact on splicing (score 0.99). Therefore, PP3 is applied at Supporting strength because computational evidence supports a deleterious splicing effect.
PP4
PP4 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP4 requires a specific phenotype with a single genetic etiology. No phenotype information is provided. Therefore, PP4 is not applied.
PP5
PP5 (Supporting)
According to standard ACMG guidelines, PP5 is: 'Reputable source reports variant as pathogenic without accessible evidence.' ClinVar lists this variant as Pathogenic from three clinical laboratories. Therefore, PP5 is applied at Supporting strength.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, BA1 applies when allele frequency >0.056%. The variant is absent from population databases. Therefore, BA1 is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines, BS1 applies for allele frequency between 0.0043% and 0.056%. The variant is absent. Therefore, BS1 is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to VCEP guidelines, BS2 requires homozygous observations in unaffected individuals. No such data exist. Therefore, BS2 is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines, BS3 requires functional studies showing no damaging effect. No functional data are available. Therefore, BS3 is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to VCEP guidelines, BS4 requires lack of segregation in affected families. No segregation data are provided. Therefore, BS4 is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP1 applies to missense variants in genes where only LOF is pathogenic. This is a splice variant causing LOF. Therefore, BP1 is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to VCEP guidelines, BP2 requires observations in trans or cis with other PTEN variants. No such data are reported. Therefore, BP2 is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP3 applies to in-frame indels in repeat regions. Not applicable to this splice variant. Therefore, BP3 is not applied.
BP4
BP4 (Not Applied) Strength Modified
According to VCEP guidelines, BP4 requires computational evidence of no impact. Computational predictions are discordant but SpliceAI indicates impact. Therefore, BP4 is not applied.
BP5
BP5 (Not Applied) Strength Modified
According to VCEP guidelines, BP5 applies when an alternate molecular basis is found. No alternate etiology is reported. Therefore, BP5 is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP6 applies to reputable sources reporting a variant as benign without evidence. ClinVar reports this variant as pathogenic, not benign. Therefore, BP6 is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to VCEP guidelines, BP7 applies to synonymous or deep intronic variants predicted to have no impact. This is a canonical splice site variant. Therefore, BP7 is not applied.