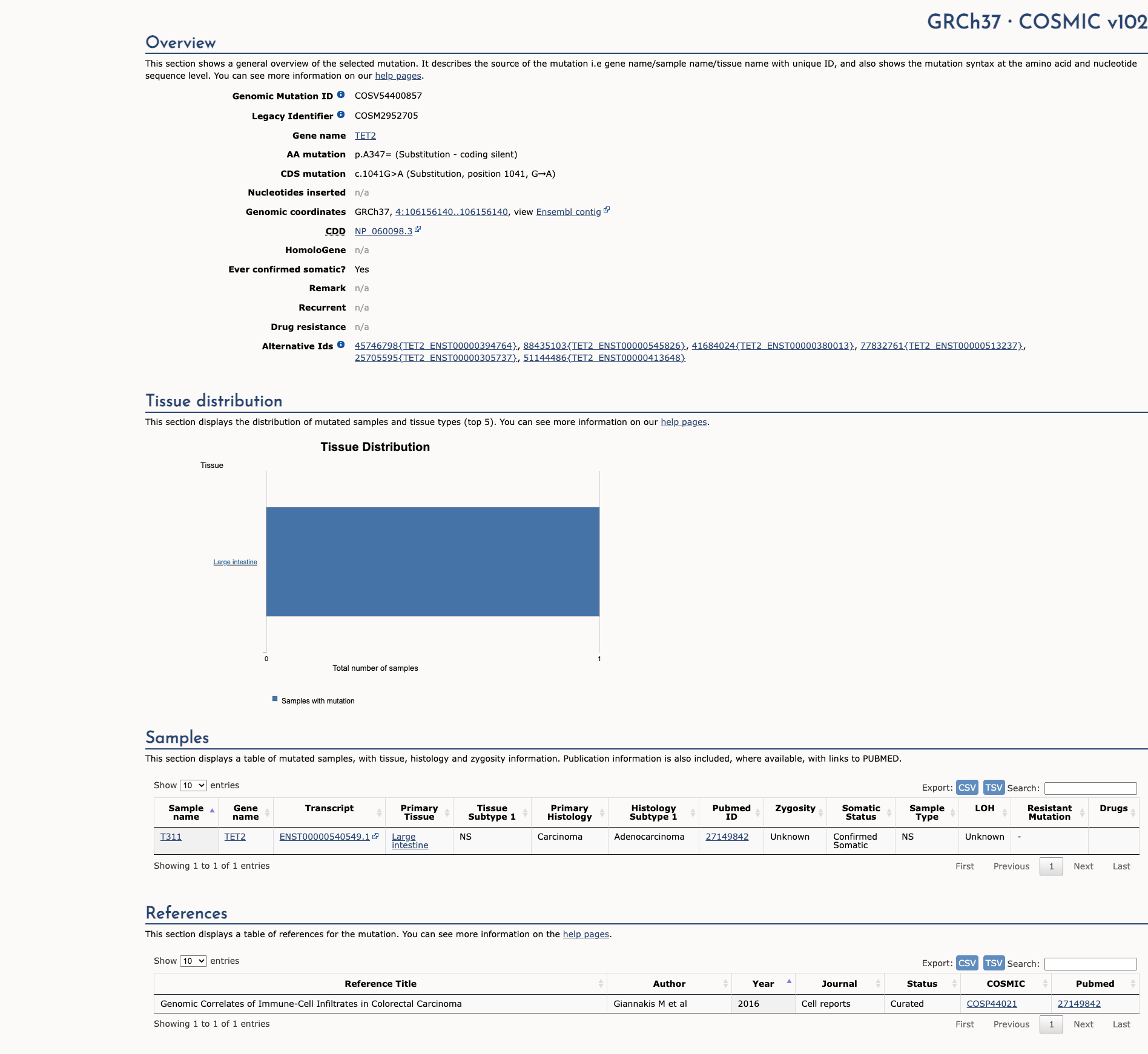
TET2 c.1041G>A, p.Ala347=
NM_001127208.2:c.1041G>A
COSMIC ID: COSM2952706
Likely Benign
This synonymous TET2 variant is absent from population databases (PM2) and computational/splice predictions indicate no impact on gene function (BP4, BP7). Additionally, reputable sources report it as benign (BP6). The combined benign evidence supports a Likely Benign classification.
ACMG/AMP Criteria Applied
PM2
BP4
BP6
BP7
Genetic Information
Gene & Transcript Details
Gene
TET2
Transcript
NM_001127208.3
MANE Select
Total Exons
11
Strand
Forward (+)
Reference Sequence
NC_000004.11
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_001127208.1 | Alternative | 11 exons | Forward |
| NM_001127208.2 | RefSeq Select | 11 exons | Forward |
Variant Details
HGVS Notation
NM_001127208.2:c.1041G>A
Protein Change
A347=
Location
Exon 3
(Exon 3 of 11)
5'Exon Structure (11 total)3'
Functional Consequence
Loss of Function
Related Variants
Alternate Identifiers
COSM2952706
Variant interpretation based on transcript NM_001127208.3
Genome Browser
Loading genome browser...
HGVS InputNM_001127208:c.1041G>A
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Population Frequency
Global Frequency
0.0 in 100,000
Extremely Rare
Global: 0.0%
0%
0.05%
0.1%
1%
5%
10%+
ACMG Criteria Applied
PM2
This variant is not present in gnomAD (PM2 criteria applies).
Classification
Likely Benign
Based on 2 submitter reviews in ClinVar
Submitter Breakdown
2 LB
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (0)
No publication details.
Clinical Statement
This variant has been reported in ClinVar as Likely benign (2 clinical laboratories).
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
Computational Analysis
Pathogenicity Predictions
Predictor Consensus
Mixed/VUS
PP3 Applied
No
Additional Predictors
Benign:
CADD: 0.20
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific)
PVS1
PVS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PVS1 is: "Null variant (nonsense, frameshift, canonical ±1 or 2 splice sites, initiation codon, single exon deletion) in a gene where loss of function is a known mechanism of disease." The evidence for this variant shows: A347= is a synonymous variant that does not alter the protein coding sequence or create a null allele. Therefore, this criterion is not applied because the variant does not meet the definition of a null variant.
PS1
PS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS1 is: "Same amino acid change as a previously established pathogenic variant regardless of nucleotide change." The evidence for this variant shows: A347= is a synonymous change and does not alter the amino acid sequence. Therefore, this criterion is not applied because the variant does not change the amino acid.
PS2
PS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS2 is: "De novo (both maternity and paternity confirmed) in a patient with the disease and no family history." The evidence for this variant shows: No de novo parental testing data are available. Therefore, this criterion is not applied due to lack of de novo evidence.
PS3
PS3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS3 is: "Well-established functional studies show a damaging effect on the gene or gene product." The evidence for this variant shows: No functional studies have been performed. Therefore, this criterion is not applied because functional impact has not been evaluated.
PS4
PS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS4 is: "Prevalence in affected individuals significantly increased compared with controls." The evidence for this variant shows: No case-control or affected individual data are available. Therefore, this criterion is not applied because statistical evidence in cases versus controls is lacking.
PM1
PM1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM1 is: "Located in a mutational hot spot and/or critical and well-established functional domain without benign variation." The evidence for this variant shows: A347= is a synonymous change outside defined functional domains. Therefore, this criterion is not applied because the variant is not located in a hotspot or critical domain.
PM2
PM2 (Moderate)
According to standard ACMG guidelines, the rule for PM2 is: "Absent from controls (or at extremely low frequency if recessive) in Exome Sequencing Project, 1000 Genomes Project, or ExAC." The evidence for this variant shows: It is not present in gnomAD or other population databases. Therefore, this criterion is applied at Moderate strength because the variant is absent from population controls.
PM3
PM3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM3 is: "Detected in trans with a pathogenic variant for recessive disorders." The evidence for this variant shows: No trans-phase data with a pathogenic allele are available. Therefore, this criterion is not applied due to lack of trans genotype information.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM4 is: "Protein length changes due to in-frame deletions/insertions or stop-loss variants." The evidence for this variant shows: A347= does not alter protein length. Therefore, this criterion is not applied because the variant does not change protein length.
PM5
PM5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM5 is: "Novel missense change at an amino acid residue where a different pathogenic missense change has been seen before." The evidence for this variant shows: It is a synonymous change, not a missense variant. Therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM6 is: "Assumed de novo, but without confirmation of paternity and maternity." The evidence for this variant shows: No parental testing data are available. Therefore, this criterion is not applied due to lack of evidence for assumed de novo occurrence.
PP1
PP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP1 is: "Co-segregation with disease in multiple affected family members in a gene definitively known to cause the disease." The evidence for this variant shows: No segregation analysis data are provided. Therefore, this criterion is not applied due to absence of family segregation data.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP2 is: "Missense variant in a gene with a low rate of benign missense variation and where missense variants are a common mechanism of disease." The evidence for this variant shows: A347= is a synonymous variant. Therefore, this criterion is not applied because it is not a missense change.
PP3
PP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP3 is: "Multiple lines of computational evidence support a deleterious effect on the gene or gene product." The evidence for this variant shows: Computational predictors (CADD 0.20, SpliceAI 0.00) do not support deleterious effect. Therefore, this criterion is not applied because in silico evidence does not indicate a damaging impact.
PP4
PP4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP4 is: "Patient's phenotype or family history is highly specific for a disease with a single genetic etiology." The evidence for this variant shows: No phenotype or clinical information is provided. Therefore, this criterion is not applied due to lack of specific phenotypic data.
PP5
PP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP5 is: "Reputable source recently reports variant as pathogenic, but the evidence is not available to the laboratory to perform an independent evaluation." The evidence for this variant shows: No reputable source reports pathogenicity; ClinVar reports likely benign. Therefore, this criterion is not applied because no pathogenic reports exist.
BA1
BA1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BA1 is: "Allele frequency is too high for the disorder." The evidence for this variant shows: Absent from population databases. Therefore, this criterion is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS1 is: "Allele frequency is greater than expected for the disorder." The evidence for this variant shows: Absent from controls. Therefore, this criterion is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS2 is: "Observed in healthy individuals with full penetrance expected at an early age." The evidence for this variant shows: No observations in healthy individuals. Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS3 is: "Well-established functional studies show no damaging effect on protein function or splicing." The evidence for this variant shows: No functional studies available. Therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS4 is: "Lack of segregation in affected members of a family." The evidence for this variant shows: No segregation data. Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP1 is: "Missense variant in a gene for which primarily truncating variants are known to cause disease." The evidence for this variant shows: This is a synonymous variant. Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP2 is: "Observed in trans with a pathogenic variant for a dominant gene or in cis with a pathogenic variant in any inheritance pattern." The evidence for this variant shows: No phase data. Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP3 is: "In-frame insertions/deletions in a repetitive region without a known function." The evidence for this variant shows: It is a nucleotide substitution. Therefore, this criterion is not applied.
BP4
BP4 (Supporting)
According to standard ACMG guidelines, the rule for BP4 is: "Multiple lines of computational evidence suggest no impact on gene or gene product (conservation, evolutionary, splicing impact, etc.)." The evidence for this variant shows: Mixed in silico predictors with SpliceAI 0.00 indicate no impact on splicing or protein function. Therefore, this criterion is applied at Supporting strength because computational evidence does not support a deleterious effect.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP5 is: "Variant found in a case with an alternate molecular basis for disease." The evidence for this variant shows: No case data with alternate molecular diagnosis. Therefore, this criterion is not applied.
BP6
BP6 (Supporting)
According to standard ACMG guidelines, the rule for BP6 is: "Reputable source reports variant as benign, but the evidence is not available to the laboratory to perform an independent evaluation." The evidence for this variant shows: ClinVar entries from two laboratories classify it as likely benign without underlying evidence. Therefore, this criterion is applied at Supporting strength because a reputable source has reported benign classification.
BP7
BP7 (Supporting)
According to standard ACMG guidelines, the rule for BP7 is: "Synonymous variant with no predicted impact on splicing." The evidence for this variant shows: A347= is synonymous and SpliceAI predicts no splicing impact. Therefore, this criterion is applied at Supporting strength because the variant is synonymous with no splicing effect.