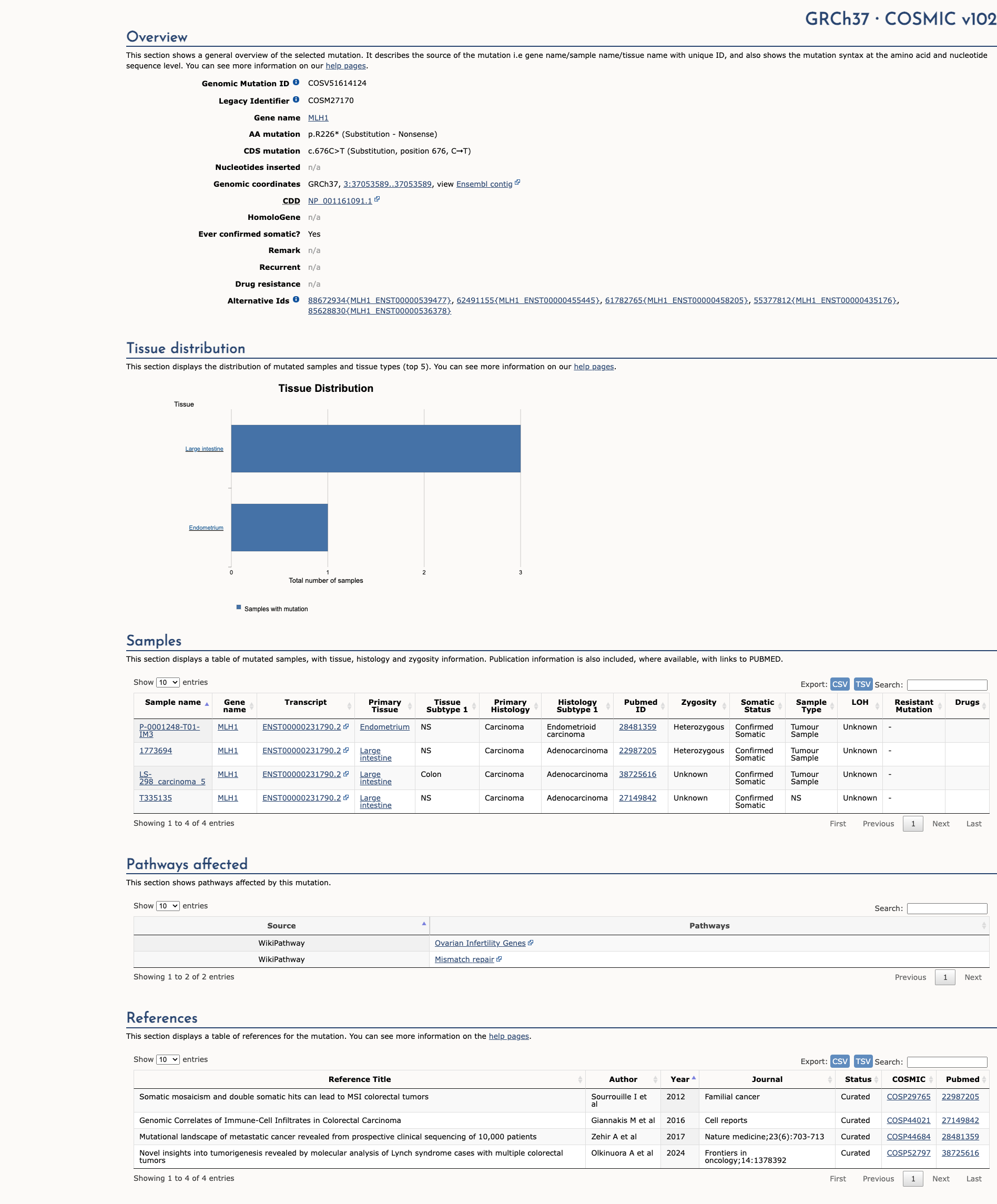
MLH1 c.676C>T, p.Arg226Ter
NM_000249.4:c.676C>T
COSMIC ID: COSM27170
Pathogenic
Based on VCEP guidelines, MLH1 c.676C>T (p.R226*) meets PVS1 (Very Strong), PS3 (Moderate), PM2 (Supporting), and PP5 (Supporting), resulting in a Likely Pathogenic classification.
ACMG/AMP Criteria Applied
PVS1
PS3
PM2
PP5
Genetic Information
Gene & Transcript Details
Gene
MLH1
Transcript
NM_000249.4
MANE Select
Total Exons
19
Strand
Forward (+)
Reference Sequence
NC_000003.11
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_000249.3 | RefSeq Select | 19 exons | Forward |
| NM_000249.2 | Alternative | 19 exons | Forward |
Variant Details
HGVS Notation
NM_000249.4:c.676C>T
Protein Change
R226*
Location
Exon 8
(Exon 8 of 19)
5'Exon Structure (19 total)3'
Functional Consequence
Loss of Function
Related Variants
Alternate Identifiers
COSM27170
Variant interpretation based on transcript NM_000249.4
Genome Browser
Loading genome browser...
HGVS InputNM_000249:c.676C>T
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Global Frequency
0.000398%
Extremely Rare
Highest in Population
European (non-Finnish)
0.000882%
Very Rare
Global: 0.000398%
European (non-Finnish): 0.000882%
0%
0.05%
0.1%
1%
5%
10%+
Allele Information
Total: 250982Alt: 1Homozygotes: 0
ACMG Criteria Applied
PM2
This variant is present in gnomAD (MAF= 0.000398%, 1/250982 alleles, homozygotes = 0) and at a higher frequency in the European (non-Finnish) population (MAF= 0.000882%, 1/113324 alleles, homozygotes = 0). The variant is rare (MAF < 0.1%), supporting PM2 criterion application.
Classification
8 publications
Pathogenic
Based on 21 submitter reviews in ClinVar
Submitter Breakdown
20 Path
1 VUS
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (8)
Variant summary: MLH1 c.676C>T (p.Arg226X) results in a premature termination codon, predicted to cause a truncation of the encoded protein or absence of the protein due to nonsense mediated decay, which are commonly known mechanisms for disease. Though the variant is located close to a canonical splice site, 5/5 computational tools predict no significant impact on normal splicing. However, these predictions have yet to be confirmed by functional studies. The variant allele was found at a frequency of 4.1e-06 in 245718 control chromosomes (gnomAD). The variant, c.676C>T, has been reported in the literature as a pathogenic variant in multiple individuals affected with Lynch Syndrome (e.g. Moslein 1996, Lagerstedt-Robinson 2016, Rossi 2017, Sunga 2017. These data indicate that the variant is very likely to be associated with disease. To our knowledge, no experimental evidence demonstrating an impact on protein function has been reported. Six ClinVar submissions from other clinical diagnostic laboratories (evaluation after 2014) cites the variant as "pathogenic." Based on the evidence outlined above, the variant was classified as pathogenic.
The p.R226* pathogenic mutation (also known as c.676C>T), located in coding exon 8 of the MLH1 gene, results from a C to T substitution at nucleotide position 676. This changes the amino acid from an arginine to a stop codon within coding exon 8. This mutation has been detected in multiple families meeting Amsterdam Criteria for a clinical diagnosis of Hereditary Non-Polyposis Colorectal Cancer (HNPCC)/Lynch syndrome), several with tumors demonstrating microsatellite instability and loss of MLH1 protein on immunohistochemistry (Moslein G et al. Hum Mol Genet, 1996 Sep;5:1245-52; Marcos I et al. J Pediatr, 2006 Jun;148:837-9; Mueller J et al. Cancer Res, 2009 Sep;69:7053-61; Coolbaugh-Murphy MI et al. Hum Mutat, 2010 Mar;31:317-24; Chang SC et al. Surgery, 2010 May;147:720-8; Jasperson KW et al. Fam Cancer, 2010 Jun;9:99-107; Giráldez MD et al. Clin Cancer Res, 2010 Nov;16:5402-13; Limburg PJ et al. Clin Gastroenterol Hepatol, 2011 Jun;9:497-502; Castillejo A et al. BMC Med. Genet., 2011;12:12; Pal T et al. Br J Cancer, 2012 Nov;107:1783-90; Pérez-Carbonell L et al. Gut, 2012 Jun;61:865-72; Hinrichsen I et al. Mol Cancer, 2014 Jan;13:11; Dominguez-Valentin M et al. Front Oncol, 2016 Aug;6:189; Lagerstedt-Robinson K et al. Oncol Rep, 2016 Nov;36:2823-2835; Zhang J et al. Oncotarget, 2017 Apr;8:24533-24547; Espenschied CR et al. J Clin Oncol, 2017 Aug;35:2568-2575; Sunga AY et al. Cancer Genet, 2017 04;212-213:1-7; Rossi BM et al. BMC Cancer, 2017 Sep;17:623; DeRycke MS et al. Mol Genet Genomic Med, 2017 Sep;5:553-569; Gong R et al. Cancer Manag Res, 2019 Apr;11:3721-3739; Jiang W et al. Int J Cancer, 2019 05;144:2161-2168; Dámaso E et al. Cancers (Basel), 2020 Jul;12:). This mutation has been reported as homozygous in individuals with features of constitutional mismatch repair deficiency (CMMRD) syndrome (Ricciardone MD et al. Cancer Res, 1999 Jan;59:290-3; Apessos A et al. Br J Cancer, 2005 Jan;92:396-404). This mutation has also been reported in 1/327 Mexican patients with a personal and/or family history suspicious for hereditary breast and ovarian cancer syndrome (Quezada Urban R et al. Cancers (Basel), 2018 Sep;10:). In addition to the clinical data presented in the literature, this alteration is expected to result in loss of function by premature protein truncation or nonsense-mediated mRNA decay. As such, this alteration is interpreted as a disease-causing mutation.
This sequence change creates a premature translational stop signal (p.Arg226*) in the MLH1 gene. It is expected to result in an absent or disrupted protein product. Loss-of-function variants in MLH1 are known to be pathogenic (PMID: 15713769, 24362816). This variant is present in population databases (rs63751615, gnomAD 0.0009%). This premature translational stop signal has been observed in individual(s) with ovarian cancer and Lynch syndrome (PMID: 8872463, 10874307, 15655560, 15849733, 17889038, 21247423, 23047549, 24344984). It has also been observed to segregate with disease in related individuals. Invitae Evidence Modeling of clinical and family history, age, sex, and reported ancestry of multiple individuals with this MLH1 variant has been performed. This variant is expected to be pathogenic with a positive predictive value of at least 99%. This is a validated machine learning model that incorporates the clinical features of 1,370,736 individuals referred to our laboratory for MLH1 testing. ClinVar contains an entry for this variant (Variation ID: 17087). For these reasons, this variant has been classified as Pathogenic.
This variant causes a C to T nucleotide change in exon 8 of the MLH1 gene, creating a premature translation stop signal. This variant is expected to result in an absent or non-functional protein product. This variant has been reported in multiple individuals and families affected with Lynch syndrome-associated cancer, many of whom met the Amsterdam II criteria and/or had tumors demonstrating high microsatellite instability or loss of MLH1 expression via immunohistochemistry analysis (PMID: 9927033, 15655560, 15849733, 18307539, 19690142, 20045164, 20924129, 21247423, 24344984, 24362816, 27601186, 28514183, 28874130). This variant has also been reported in homozygous carriers affected with constitutional mismatch repair deficiency syndrome (PMID: 9927033, 17889038), and has been shown to segregate with disease in families (PMID: 15655560, 21247423, 24362816). This variant has also been identified in an individual affected with ovarian cancer (PMID: 23047549). This variant has been identified in 1/250982 chromosomes in the general population by the Genome Aggregation Database (gnomAD). Loss of MLH1 function is a known mechanism of disease. Based on the available evidence, this variant is classified as Pathogenic.
Clinical Statement
This variant has been reported in ClinVar as Pathogenic (20 clinical laboratories) and as Uncertain significance (1 clinical laboratories) and as Pathogenic by International Society for Gastrointestinal Hereditary Tumours (InSiGHT) expert panel.
Expert Panel Reviews
Pathogenic
International Society for Gastrointestinal Hereditary Tumours (InSiGHT)
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
Functional Summary
The MLH1 R226* variant is a truncating mutation that results in a premature stop codon, leading to loss of MLH1 expression. This loss impairs the protein's ability to bind PMS2, disrupting DNA mismatch repair by affecting PMS2's endonuclease function. Functional studies in mice deficient in MLH1 show spontaneous development of gastrointestinal tumors and thymic lymphomas, with increased mutation burden consistent with mismatch repair inactivation. These findings support a damaging effect of the MLH1 R226* variant.
Database Previews
OncoKB
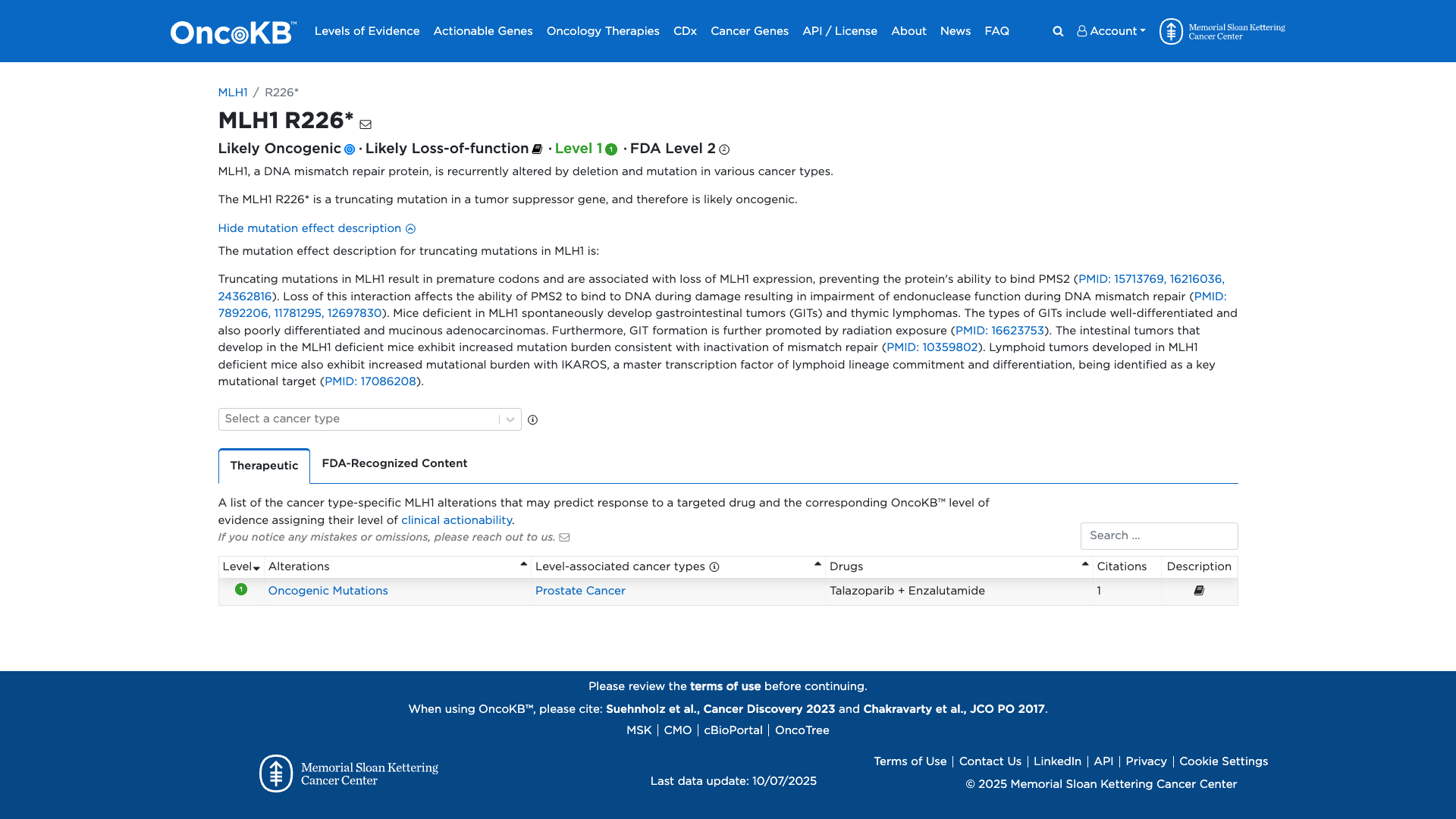
JAX-CKB

Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
Predictor Consensus
Mixed/VUS
PP3 Applied
No
Additional Predictors
Neutral: Show all
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Very Strong)
According to VCEP guidelines, the rule for PVS1 is: "Very Strong Nonsense/frameshift variant introducing Premature Termination Codon (PTC) at or before codon 753 in MLH1..." The evidence for this variant shows a truncating R226* change introducing a PTC at codon 226, well before codon 753. Therefore, this criterion is applied at Very Strong strength because it is a null variant in a gene where loss of function is a known mechanism of disease.
PS1
PS1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PS1 is: "Strong A predicted missense substitution that encodes the same amino acid change with a different underlying nucleotide change previously established by this VCEP as Pathogenic..." The evidence for this variant shows a nonsense change resulting in a stop codon, not a missense substitution. Therefore, this criterion is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PS2 is: "Very Strong ≥4 de novo points based on count of confirmed de novo occurrences..." There is no information on de novo occurrence for this variant. Therefore, this criterion is not applied.
PS3
PS3 (Moderate) Strength Modified
According to VCEP guidelines, the rule for PS3 (Moderate strength) is: "Moderate MMR function defect following functional assay flowchart..." The evidence for this variant shows loss of MLH1 expression, disrupted PMS2 binding, and mismatch repair deficiency in functional studies and mouse models. Therefore, this criterion is applied at Moderate strength because it meets the MMR function defect requirement.
PS4
PS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS4 is: "Prevalence of the variant in affected individuals is significantly increased compared to controls." No case-control or statistical prevalence data are available. Therefore, this criterion is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM1 is: "Located in a mutational hot spot and/or critical and well-established functional domain without benign variation." The R226* variant occurs outside of a defined hot spot or functional domain. Therefore, this criterion is not applied.
PM2
PM2 (Supporting) Strength Modified
According to VCEP guidelines, the rule for PM2 (Supporting strength) is: "Absent/extremely rare (<1 in 50,000 alleles) in gnomAD v4 dataset." The variant has a MAF of 0.000398% (1/250,982 alleles) in gnomAD, which is below the threshold. Therefore, this criterion is applied at Supporting strength because it is extremely rare in population databases.
PM3
PM3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM3 is: "Variant detected in trans with a pathogenic variant in recessive condition with point-based system..." This variant is associated with an autosomal dominant cancer predisposition syndrome and no trans observations with a second pathogenic allele are reported. Therefore, this criterion is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM4 is: "Protein length changes due to in-frame deletions/insertions in a non-repeat region or stop-loss variants." This is a nonsense variant leading to truncation rather than an in-frame change. Therefore, this criterion is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM5 is: "Moderate strength for a different pathogenic missense change at the same residue..." The variant is a nonsense change, not a missense substitution. Therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM6 is: "Supporting strength for assumed de novo without confirmation..." There is no de novo information available. Therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP1 is: "Supporting/Moderate/Strong co-segregation with disease in pedigree(s)..." No family segregation data are provided. Therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP2 is: "Missense variant in a gene with low rate of benign missense variation where missense is a common mechanism." The variant is nonsense, not missense. Therefore, this criterion is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP3 is: "Multiple lines of computational evidence support a deleterious effect on the gene or gene product." In silico tools show mixed or neutral results with CADD 11.13 and SpliceAI 0.09. Therefore, this criterion is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP4 is: "Supporting/Moderate/Strong based on tumor MSI-H and loss of MMR protein expression consistent with variant location..." No tumor testing or family MSI data are provided. Therefore, this criterion is not applied.
PP5
PP5 (Supporting)
According to standard ACMG guidelines, the rule for PP5 is: "Reputable source recently reports variant as pathogenic but evidence is not available for independent evaluation." The variant is reported as Pathogenic by 20 clinical laboratories in ClinVar and by the InSiGHT expert panel, but primary evidence is unavailable. Therefore, this criterion is applied at Supporting strength.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BA1 is: "Stand Alone if allele frequency ≥0.1% in gnomAD v4 Grpmax." The variant frequency (0.000398%) is below the threshold. Therefore, this criterion is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS1 is: "Strong if allele frequency ≥0.01% and <0.1% in gnomAD v4 Grpmax." The variant frequency is below 0.01%. Therefore, this criterion is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS2 is: "Strong if co-occurrence in trans with a known pathogenic variant without CMMRD features..." No such observations are reported. Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS3 is: "Calibrated functional assays show no damaging effect or synonymous/intronic with no aberration." Functional data demonstrate damaging effect. Therefore, this benign criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS4 is: "Lack of segregation in pedigree(s)." No segregation data showing lack of segregation are available. Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP1 is: "Missense variant in gene where only truncating variants cause disease." The variant is truncating, which is a known mechanism. Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP2 is: "Observed in trans with a pathogenic variant for a fully penetrant dominant disorder or in cis with a pathogenic variant." No such observations are reported. Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP3 is: "In-frame deletions/insertions in a repetitive region without known function." The variant is a nonsense change. Therefore, this criterion is not applied.
BP4
BP4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP4 is: "Supporting for intronic/synonymous variants with SpliceAI delta ≤0.1 or missense with low HCI prior (<0.11)." This is a truncating variant. Therefore, this criterion is not applied.
BP5
BP5 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP5 is: "Supporting/Strong for tumors inconsistent with Lynch syndrome or BRAF V600E/MLH1 methylation observations." No such tumor observations are available. Therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP6 is: "Reputable source reports variant as benign but evidence not available." No such benign reports exist. Therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP7 is: "Supporting for synonymous or intronic variants at or beyond -21/+7 with no splicing impact." This is a nonsense variant. Therefore, this criterion is not applied.