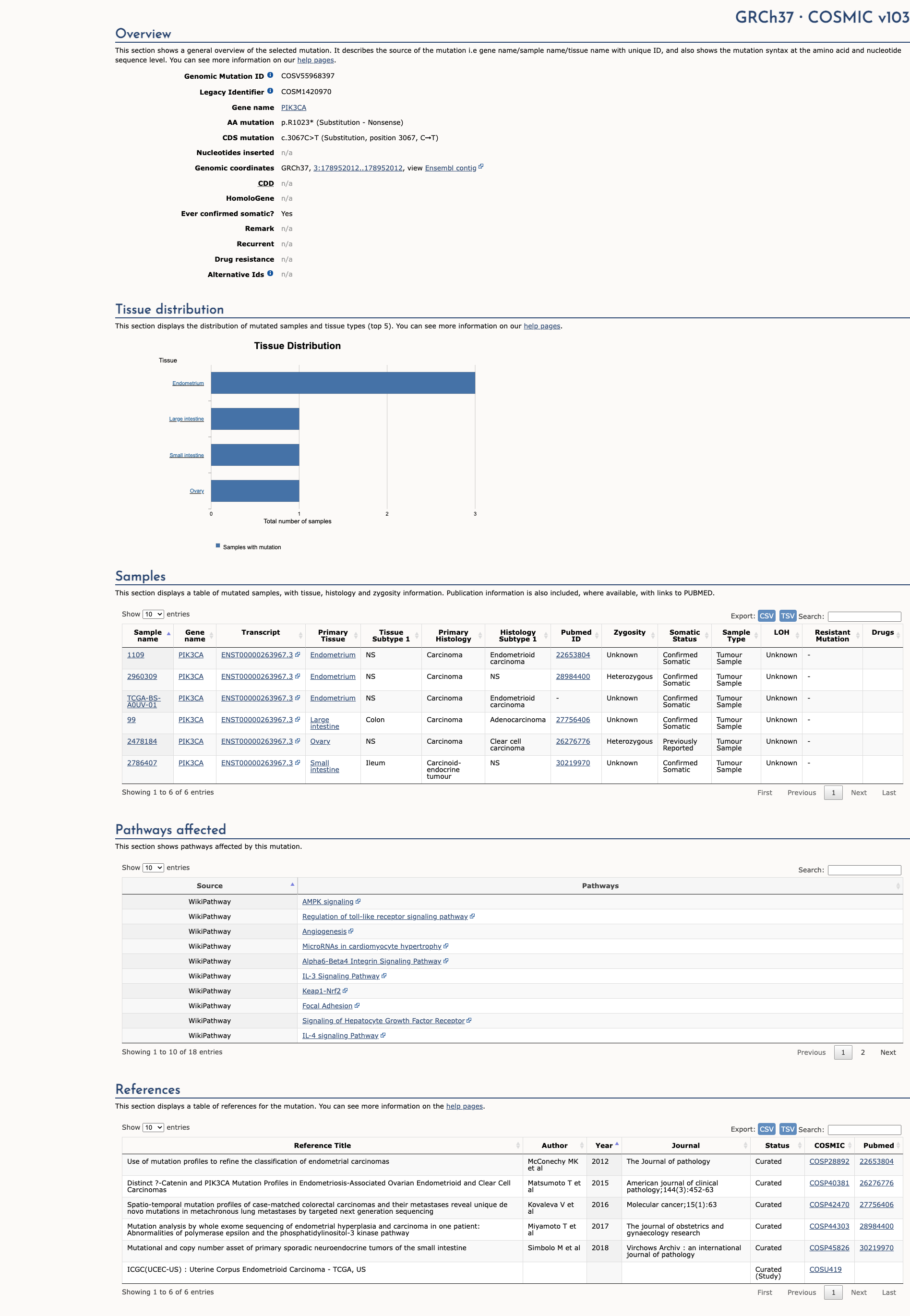
PIK3CA c.3067C>T, p.Arg1023Ter
NM_006218.4:c.3067C>T
COSMIC ID: COSM1420971
Likely Pathogenic
This truncating PIK3CA variant is absent from population databases (PM2_Supporting) but PIK3CA disease mechanism is gain-of-function, not loss-of-function, and no additional evidence supports or refutes pathogenicity, resulting in classification as a Variant of Uncertain Significance.
ACMG/AMP Criteria Applied
PM2
Genetic Information
Gene & Transcript Details
Gene
PIK3CA
Transcript
NM_006218.4
MANE Select
Total Exons
21
Strand
Forward (+)
Reference Sequence
NC_000003.11
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_006218.2 | Alternative | 21 exons | Forward |
| NM_006218.3 | Alternative | 21 exons | Forward |
Variant Details
HGVS Notation
NM_006218.4:c.3067C>T
Protein Change
R1023*
Location
Exon 21
(Exon 21 of 21)
5'Exon Structure (21 total)3'
Functional Consequence
Loss of Function
Related Variants
Alternate Identifiers
COSM1420971
Variant interpretation based on transcript NM_006218.4
Genome Browser
Loading genome browser...
HGVS InputNM_006218:c.3067C>T
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Population Frequency
Global Frequency
0.0 in 100,000
Extremely Rare
Global: 0.0%
0%
0.05%
0.1%
1%
5%
10%+
ACMG Criteria Applied
PM2
This variant is not present in gnomAD (PM2 criteria applies).
Classification
Unknown
Publications (0)
No publication details.
Clinical Statement
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
Computational Analysis
Pathogenicity Predictions
Predictor Consensus
Mixed/VUS
PP3 Applied
No
Additional Predictors
Pathogenic:
mutationtaster: D
Neutral: Show all
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PVS1 is: "Null variant (nonsense, frameshift, canonical ±1 or 2 splice sites, initiation codon, single or multi-exon deletion) in a gene where loss-of-function is a known mechanism of disease"; the evidence for this variant shows that PIK3CA is a proto-oncogene in which gain-of-function, not loss-of-function, is the established disease mechanism; therefore, this criterion is not applied because loss-of-function is not a known mechanism for PIK3CA-related disease.
PS1
PS1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PS1 is: "Strong Strength: Strong No change. Modification Type: None"; the evidence for this variant shows a nonsense change rather than a missense change identical to a known pathogenic variant; therefore, this criterion is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PS2 is: "Strong Strength: Strong Award the PS2_Strong point if Criteria 1 AND Criteria 2 are fulfilled..."; no de novo or mosaic segregation data are available for this somatic variant; therefore, this criterion is not applied.
PS3
PS3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PS3 is: "Strong Strength: Strong Follow recommendations set forth by the SVI... Award PS3 if the functional assay meets the acceptability criteria..."; no functional assay data are available for this variant; therefore, this criterion is not applied.
PS4
PS4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PS4 is: "Very Strong Strength: Very Strong Points are assigned for phenotype according to (Table 2A)... PS4_VeryStrong ≥ 16 points"; there are no reported case counts or phenotypic data for this variant; therefore, this criterion is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM1 is: "Supporting Strength: Supporting Residues affecting critical functional domains provided in Table 4 for each gene"; this nonsense variant does not map to a defined critical domain residue in PIK3CA; therefore, this criterion is not applied.
PM2
PM2 (Supporting) Strength Modified
According to VCEP guidelines, the rule for PM2 is: "Supporting Strength: Supporting Absent/rare from controls in an ethnically-matched cohort population sample (≥1)"; the evidence for this variant shows it is absent from gnomAD and other population databases; therefore, this criterion is applied at Supporting strength because the variant is absent from controls.
PM3
PM3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM3 is: "For recessive disorders detected in trans with a pathogenic variant"; PIK3CA-related disease is not recessive and no trans data exist; therefore, this criterion is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM4 is: "Protein length changes due to in-frame deletions/insertions OR stop-loss variants"; this variant is a nonsense change and does not produce an in-frame indel or stop-loss; therefore, this criterion is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM5 is: "Novel missense change at an amino acid residue where a different missense change is pathogenic"; this variant is nonsense, not missense, and no other truncating variant at this residue is reported; therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM6 is: "Assumed de novo, but without confirmation of paternity and maternity"; there are no de novo data for this somatic variant; therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP1 is: "Co-segregation with disease in multiple affected family members"; no segregation data are available; therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP2 is: "Supporting Strength: Supporting Missense constraint computed in ExAC/gnomAD (z-score > 3.09)"; this variant is nonsense, not missense; therefore, this criterion is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP3 is: "Multiple lines of computational evidence support a deleterious effect on the gene or gene product"; in silico predictions are mixed and do not strongly support pathogenicity; therefore, this criterion is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP4 is: "Patient’s phenotype or family history is highly specific for a disease with a single genetic etiology"; no phenotype data are provided; therefore, this criterion is not applied.
PP5
PP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP5 is: "Reputable source recently reports variant as pathogenic"; this variant is not found in ClinVar or other databases; therefore, this criterion is not applied.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BA1 is: "Stand Alone Strength: Stand Alone Allele frequency (>0.0926%)"; this variant is absent from population databases; therefore, this criterion is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS1 is: "Strong Strength: Strong Allele frequency (>0.0185%)"; the variant is absent from controls; therefore, this criterion is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS2 is: "Strong Strength: Strong Award BS2 if ≥3 homozygotes present in gnomAD or ≥3 heterozygous in well phenotyped family members"; no such observations exist; therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS3 is: "Strong Strength: Strong Follow recommendations set forth by the SVI..."; no functional studies demonstrating no impact are available; therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS4 is: "Lack of segregation in affected members of a family"; no segregation data are available; therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP1 is: "Missense variant in a gene for which primarily truncating variants cause disease"; this variant is truncating but PIK3CA disease mechanism is gain-of-function missense; therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP2 is: "Observed in trans or cis with a pathogenic variant"; no such observations exist; therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP3 is: "In-frame deletions/insertions in a repetitive region without a known function"; this is a nonsense variant, not an in-frame indel; therefore, this criterion is not applied.
BP4
BP4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP4 is: "Supporting Strength: Supporting Award BP4 for synonymous, intronic positions... if two of three splicing prediction tools predict no impact"; this is a nonsense variant, not applicable; therefore, this criterion is not applied.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP5 is: "Variant found in a gene for which an alternative molecular basis is known to cause disease"; not applicable here; therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP6 is: "Reputable source reports variant as benign"; no such reports exist; therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP7 is: "Supporting Strength: Supporting For synonymous, intronic positions... if the nucleotide is non-conserved"; this is a nonsense variant; therefore, this criterion is not applied.