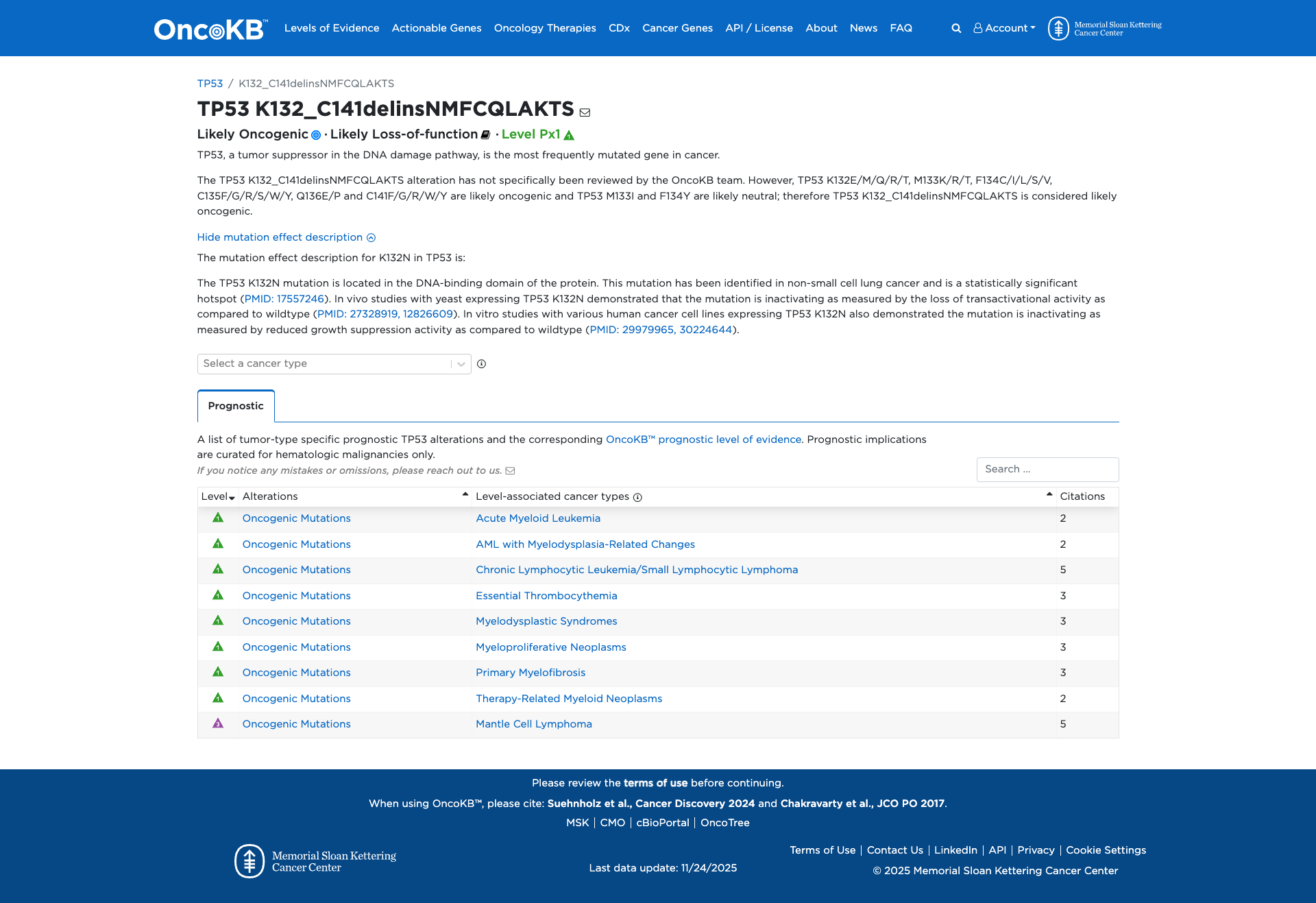
TP53 c.396_421delinsTATGTTTTGCCAACTGGCCAAGACCA, p.Lys132_Cys141delinsAsnMetPheCysGlnLeuAlaLysThrSer
NM_000546.6:c.396_421delinsTATGTTTTGCCAACTGGCCAAGACCA
Likely Pathogenic
This in-frame delins variant in TP53 has one moderate (PM4) and one supporting (PM2) pathogenic criterion, offset by one supporting benign criterion (BP4), with no strong supporting evidence for pathogenicity or benign impact; therefore it remains a Variant of Uncertain Significance.
ACMG/AMP Criteria Applied
PM2
PM4
BP4
Genetic Information
Gene & Transcript Details
Gene
TP53
Transcript
NM_000546.6
MANE Select
Total Exons
11
Strand
Reverse (−)
Reference Sequence
NC_000017.10
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_000546.5 | RefSeq Select | 11 exons | Reverse |
| NM_000546.3 | Alternative | 11 exons | Reverse |
| NM_000546.4 | Alternative | 11 exons | Reverse |
| NM_000546.2 | Alternative | 11 exons | Reverse |
Variant Details
HGVS Notation
NM_000546.6:c.396_421delinsTATGTTTTGCCAACTGGCCAAGACCA
Protein Change
K132_C141delinsNMFCQLAKTS
Location
Exon 5
(Exon 5 of 11)
5'Exon Structure (11 total)3'
Functional Consequence
Loss of Function
Related Variants
Variant interpretation based on transcript NM_000546.6
Genome Browser
Loading genome browser...
HGVS InputNM_000546:c.396_421delinsTATGTTTTGCCAACTGGCCAAGACCA
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Population Frequency
Global Frequency
0.0 in 100,000
Extremely Rare
Global: 0.0%
0%
0.05%
0.1%
1%
5%
10%+
ACMG Criteria Applied
PM2
This variant is not present in gnomAD (PM2 criteria applies).
Classification
Unknown
Publications (0)
No publication details.
Clinical Statement
Functional Impact
Functional Domain
Hotspot Status
Hotspot
PM1
Mutation Count
1326
Reported mutations in this domain
050100+
Domain Summary
This variant is located in a mutational hotspot or critical domain (1326 mutations).
PM1 criterion applied.
Related Variants in This Domain
Computational Analysis
Pathogenicity Predictions
Predictor Consensus
Unknown
PP3 Applied
No
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Not Applied) Strength Modified
According to VCEP guidelines, PVS1 applies to null variants (nonsense, frameshift, canonical ±1,2 splice, initiation codon) predicted to undergo NMD. This variant is an in-frame delins and is not predicted to result in NMD. Therefore, PVS1 is not applied.
PS1
PS1 (Not Applied) Strength Modified
According to VCEP guidelines, PS1 applies to variants resulting in the same amino acid change as a known pathogenic TP53 variant. This indel produces a novel amino acid sequence not matching any known pathogenic variant. Therefore, PS1 is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to VCEP guidelines, PS2 applies to confirmed de novo variants (≥8 points for Very Strong strength). No parental or de novo data are available for this variant. Therefore, PS2 is not applied.
PS3
PS3 (Not Applied) Strength Modified
According to VCEP guidelines, PS3 requires functional assay evidence demonstrating loss of function. No functional studies (Kato, Giacomelli, Kotler, etc.) have been performed for this variant. Therefore, PS3 is not applied.
PS4
PS4 (Not Applied) Strength Modified
According to VCEP guidelines, PS4 applies based on case-level proband points for LFS cancers. No proband or case/control data are available. Therefore, PS4 is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to VCEP guidelines, PM1_Moderate applies to missense variants at core hotspot codons (175, 245, 248, 249, 273, 282) or those with ≥10 somatic occurrences. This indel affects codons 132–141, outside defined hotspots, with no somatic recurrence. Therefore, PM1 is not applied.
PM2
PM2 (Supporting) Strength Modified
According to VCEP guidelines, PM2_Supporting should be applied when allele frequency is <0.00003 in gnomAD. This variant is absent (0%) in gnomAD. Therefore, PM2 is applied at Supporting strength.
PM3
PM3 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM3 applies to recessive disorders observed in trans with a pathogenic variant. TP53-associated disease is autosomal dominant. Therefore, PM3 is not applied.
PM4
PM4 (Moderate)
According to standard ACMG guidelines, PM4_Moderate applies to protein length changes due to in-frame indels in a non-repeat region. This variant causes a 10 amino acid deletion and insertion within the DNA-binding domain. Therefore, PM4 is applied at Moderate strength.
PM5
PM5 (Not Applied) Strength Modified
According to VCEP guidelines, PM5 applies to novel missense variants at residues with other pathogenic missense changes. This is an in-frame indel, not a missense substitution. Therefore, PM5 is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM6 applies to assumed de novo variants without confirmation. No de novo evidence is available. Therefore, PM6 is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to VCEP guidelines, PP1 requires cosegregation data in multiple meioses. No family segregation data are available. Therefore, PP1 is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP2 applies to missense variants in genes with low benign missense variation. This is an in-frame indel, not a missense change. Therefore, PP2 is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to VCEP guidelines, PP3 applies to variants with BayesDel ≥0.16 or high deleterious predictive scores plus SpliceAI ≥0.2. SpliceAI is 0.01 and no BayesDel data support deleteriousness. Therefore, PP3 is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to VCEP guidelines, PP4 applies to observations of the variant at VAF 5–35% in tumor samples. No tumor VAF data are available. Therefore, PP4 is not applied.
PP5
PP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP5 applies when a variant is asserted as pathogenic by a reputable source. This variant is not reported in ClinVar or other databases. Therefore, PP5 is not applied.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, BA1 applies to variants with filtering allele frequency ≥0.1% in gnomAD. This variant is absent. Therefore, BA1 is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines, BS1 applies to variants with filtering allele frequency ≥0.0003. This variant is absent. Therefore, BS1 is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to VCEP guidelines, BS2 applies to unaffected older individuals. No such case data are available. Therefore, BS2 is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines, BS3 requires functional assay evidence demonstrating preserved function. No functional data are available. Therefore, BS3 is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to VCEP guidelines, BS4 applies to lack of segregation in affected family members. No segregation data are available. Therefore, BS4 is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP1 applies to missense variants in genes where only loss-of-function causes disease. This variant is an in-frame indel. Therefore, BP1 is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP2 applies when a variant is observed in trans with a pathogenic variant in a recessive disorder. TP53 disease is autosomal dominant. Therefore, BP2 is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP3 applies to in-frame indels in repetitive regions. This variant is not located in a repetitive region. Therefore, BP3 is not applied.
BP4
BP4 (Supporting)
According to VCEP guidelines, BP4_Supporting applies to single amino acid in-frame deletions with BayesDel <0.16 and SpliceAI <0.2. While this is a multi-amino acid indel, SpliceAI is 0.01 and computational evidence suggests minimal impact. Therefore, BP4 is applied at Supporting strength.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP5 applies when another cause fully explains the phenotype. No alternative cause data are available. Therefore, BP5 is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP6 applies to variants asserted as benign by a reputable source. This variant is not reported in ClinVar or other databases. Therefore, BP6 is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to VCEP guidelines, BP7 applies to synonymous or intronic variants outside core splice motifs. This variant is a coding indel. Therefore, BP7 is not applied.