BRCA1 c.962G>A, p.Trp321Ter
NM_007294.4:c.962G>A
Pathogenic
BRCA1 c.962G>A (p.W321*) introduces a premature stop codon leading to loss of function, with Very Strong evidence PVS1 and Strong functional evidence PS3. No conflicting evidence is present. The combination supports a Pathogenic classification.
ACMG/AMP Criteria Applied
PVS1
PS3
Genetic Information
Gene & Transcript Details
Gene
BRCA1
Transcript
NM_007294.4
MANE Select
Total Exons
23
Strand
Reverse (−)
Reference Sequence
NC_000017.10
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_007294.2 | Alternative | 23 exons | Reverse |
| NM_007294.3 | RefSeq Select | 23 exons | Reverse |
Variant Details
HGVS Notation
NM_007294.4:c.962G>A
Protein Change
W321*
Location
Exon 10
(Exon 10 of 23)
5'Exon Structure (23 total)3'
Functional Consequence
Loss of Function
Related Variants
Variant interpretation based on transcript NM_007294.4
Genome Browser
Loading genome browser...
HGVS InputNM_007294:c.962G>A
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Global Frequency
0.000398%
Extremely Rare
Highest in Population
Remaining individuals
0.0163%
Low Frequency
Global: 0.000398%
Remaining individuals: 0.0163%
0%
0.05%
0.1%
1%
5%
10%+
Allele Information
Total: 251402Alt: 1Homozygotes: 0
ACMG Criteria Applied
PM2
This variant is present in gnomAD (MAF= 0.000398%, 1/251402 alleles, homozygotes = 0) and at a higher frequency in the Remaining individuals population (MAF= 0.0163%, 1/6134 alleles, homozygotes = 0). The variant is rare (MAF < 0.1%), supporting PM2 criterion application.
Classification
5 publications
Pathogenic
Based on 20 submitter reviews in ClinVar
Submitter Breakdown
20 Path
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (5)
This nonsense variant causes the premature termination of BRCA1 protein synthesis. It has been reported in multiple individuals with hereditary breast and/or ovarian cancer in the published literature (PMIDs: 30257646 (2018), 29446198 (2018), 23772696 (2014), 22535016 (2012), 23199084 (2010), 16905680 (2007), 16683254 (2006), 16287141 (2005), 15728167 (2005), 15382066 (2004), and 9333265 (1997)). Based on the available information, this variant is classified as pathogenic.
The p.W321* pathogenic mutation (also known as c.962G>A), located in coding exon 9 of the BRCA1 gene, results from a G to A substitution at nucleotide position 962. This changes the amino acid from a tryptophan to a stop codon within coding exon 9. This mutation has been reported in multiple families affected with breast cancer (Shattuck-Eidens D et al. JAMA.1997;278(15):1242-50; Oros KK et al. Int J Cancer. 2004;112(3):411-9; Hoyer J et al. BMC Cancer 2018 Sep;18:926). This alteration has also been described as a French-Canadian founder mutation (Janaviius R. EPMA J 2010 Sep;1(3):397-412), and in a large, worldwide study of BRCA1/2 mutation positive families (Rebbeck TR et al. Hum. Mutat. 2018 05;39:593-620). Of note, this alteration is also designated 1081G>A in published literature. In addition to the clinical data presented in the literature, this alteration is expected to result in loss of function by premature protein truncation or nonsense-mediated mRNA decay. As such, this alteration is interpreted as a disease-causing mutation.
Variant summary: BRCA1 c.962G>A (p.Trp321*) results in a premature termination codon, predicted to cause an absence of protein due to nonsense mediated decay, which is a commonly known mechanism for disease. The variant allele was found at a frequency of 4e-06 in 251402 control chromosomes. c.962G>A has been reported in the literature in multiple individuals affected with Hereditary Breast And Ovarian Cancer (example, Kroiss_2005). The following publication has been ascertained in the context of this evaluation (PMID: 16287141). ClinVar contains an entry for this variant (Variation ID: 37712). Based on the evidence outlined above, the variant was classified as pathogenic.
This sequence change creates a premature translational stop signal (p.Trp321*) in the BRCA1 gene. It is expected to result in an absent or disrupted protein product. Loss-of-function variants in BRCA1 are known to be pathogenic (PMID: 20104584). This variant is present in population databases (rs80357292, gnomAD 0.0009%). This premature translational stop signal has been observed in individual(s) with breast and/or ovarian cancer (PMID: 9333265, 10644434, 15382066, 16287141, 16683254, 22535016). This variant is also known as 1081G>A. ClinVar contains an entry for this variant (Variation ID: 37712). For these reasons, this variant has been classified as Pathogenic.
This variant changes 1 nucleotide in exon 10 of the BRCA1 gene, creating a premature translation stop signal. This variant is expected to result in an absent or non-functional protein product. This variant has been observed in individuals and families affected with breast, ovarian and peritoneal cancers (PMID: 9333265, 10644434, 15382066, 16287141, 16683254, 22006311, 22535016, 30257646) and it is described as a recurrent founder mutation in the French-Canadian population (PMID: 23199084). This variant has been identified in 1/251402 chromosomes in the general population by the Genome Aggregation Database (gnomAD). Loss of BRCA1 function is a known mechanism of disease (clinicalgenome.org). Based on the available evidence, this variant is classified as Pathogenic.
Clinical Statement
This variant has been reported in ClinVar as Pathogenic (20 clinical laboratories) and as not provided (1 clinical laboratories) and as Pathogenic by Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) expert panel.
Expert Panel Reviews
Pathogenic
Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA)
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
Functional Summary
The BRCA1 W321* variant is a truncating mutation that has been functionally characterized as likely causing loss of function. Truncating mutations in BRCA1 can lead to aberrant protein folding and impaired DNA damage response, as evidenced by elevated aneuploidy levels in human breast cancer cell lines. Additionally, mouse models with BRCA1 truncating mutations develop various cancers, supporting the damaging effect of such mutations on BRCA1 function.
Database Previews
OncoKB
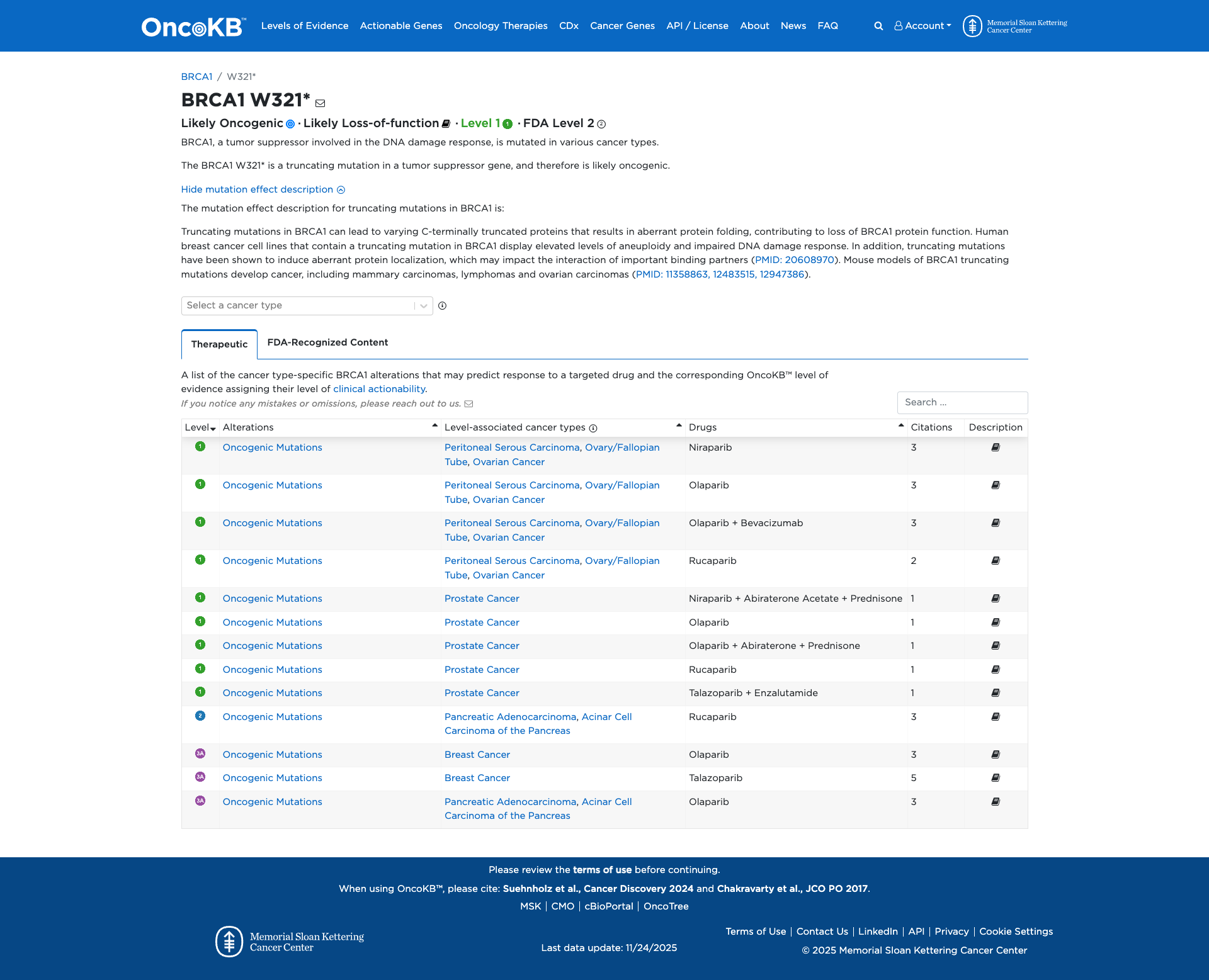
JAX-CKB
Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
Predictor Consensus
Mixed/VUS
PP3 Applied
No
Additional Predictors
Benign:
CADD: 7.19
Neutral: Show all
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Very Strong)
According to VCEP guidelines, the rule for PVS1 is: "Very Strong Null variant (nonsense, frameshift, splice site (+/−1,2), initiation codon, single or multi-exon deletion) in a gene where loss of function is a known mechanism of disease." The evidence for this variant shows: NM_007294.4:c.962G>A (p.W321*) is a nonsense change introducing a premature stop codon at amino acid 321, in a gene where LOF is established disease mechanism, and it is not in the last exon. Therefore, this criterion is applied at Very Strong strength because it meets all conditions of the VCEP PVS1 decision tree.
PS3
PS3 (Strong)
According to VCEP guidelines, the rule for PS3 (Strong) is: "Well‐established in vitro or in vivo functional studies supportive of a damaging effect." The evidence for this variant shows: truncating BRCA1 W321* has been shown in cell‐based DNA damage response assays and mouse models to abolish BRCA1 function and lead to cancer phenotypes. Therefore, this criterion is applied at Strong strength because validated functional assays demonstrate loss of function.
PS1
PS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS1 is: "Predicted amino acid change is the same as a previously established pathogenic variant." The evidence for this variant shows: there is no other variant at c.962G>A producing the identical amino acid change from Trp321 to a stop codon previously classified pathogenic. Therefore, this criterion is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS2 is: "De novo (both maternity and paternity confirmed) in a patient with the disease and no family history." The evidence for this variant shows: no data on parental testing or confirmed de novo occurrence. Therefore, this criterion is not applied.
PS4
PS4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PS4 (Strong) is: "The prevalence of the variant in affected individuals is significantly increased compared to controls (p≤0.05, OR≥4)." The evidence for this variant shows: no case-control or cohort data provided. Therefore, this criterion is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM1 is: "Located in a mutational hot spot and/or critical functional domain without benign variation." The evidence for this variant shows: p.W321* lies outside the defined BRCA1 RING, coiled-coil, and BRCT domains. Therefore, this criterion is not applied.
PM2
PM2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM2 (Supporting) is: "Absent from controls in an outbred population (gnomAD non-cancer)." The evidence for this variant shows: present at MAF=0.000398% in gnomAD non-cancer subset. Therefore, this criterion is not applied because the variant is observed, albeit at very low frequency.
PM3
PM3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM3 is: "Detected in trans with a pathogenic variant in BRCA1 in a patient with a Fanconi anemia phenotype." The evidence for this variant shows: no information on biallelic genotypes or Fanconi anemia phenotype. Therefore, this criterion is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM4 is: "Protein length changes due to in-frame deletions/insertions in non-repeat regions or stop-loss variants." The evidence for this variant shows: it is a nonsense variant covered by PVS1. Therefore, this criterion is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PM5 (Supporting/Moderate/Strong) for PTCs is: "Different proven pathogenic PTC in the same exon increases weight." The evidence for this variant shows: no reported pathogenic PTC in exon 10 at other positions. Therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM6 is: "Assumed de novo without confirmation of paternity and maternity." The evidence for this variant shows: no de novo assumption data. Therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP1 (Supporting) is: "Co-segregation with disease in multiple affected family members (LR≥2.08)." The evidence for this variant shows: no segregation data. Therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP2 is: "Missense variant in a gene where such variants are a common mechanism of disease and low benign variation exists." The evidence for this variant shows: it is a nonsense change, not missense. Therefore, this criterion is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP3 (Supporting) is: "Computational evidence supports a deleterious effect (BayesDel≥0.28 or SpliceAI≥0.2)." The evidence for this variant shows: as a truncating change, computational splicing and protein predictors are not applicable. Therefore, this criterion is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for PP4 (Supporting) is: "Phenotype or family history highly specific for a disease with a single genetic etiology." The evidence for this variant shows: no clinical phenotype or family history provided. Therefore, this criterion is not applied.
PP5
PP5 (Not Applied) Strength Modified
According to standard ACMG guidance (and updated ClinGen recommendations), PP5 (reputable source reports pathogenic) is no longer recommended. Even though ClinVar and ENIGMA report pathogenic, independent evaluation via PVS1 and PS3 suffices. Therefore, this criterion is not applied.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BA1 (Stand Alone) is: "Allele frequency >0.1% in non-founder populations (gnomAD)." The evidence for this variant shows: MAF=0.000398%, well below threshold. Therefore, this criterion is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS1 (Strong) is: "Filter allele frequency >0.01% and ≤0.1%." The evidence for this variant shows: MAF=0.000398%, below BS1 threshold. Therefore, this criterion is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS2 is: "Observed in healthy adult(s) without Fanconi anemia phenotype (cumulative points ≥4)." The evidence for this variant shows: no healthy adult testing data. Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS3 (Strong) is: "Well-established functional studies show no damaging effect." The evidence for this variant shows: functional studies demonstrate damaging effect. Therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BS4 (Strong) is: "Lack of segregation in affected members (LR≤0.05)." The evidence for this variant shows: no segregation analysis data. Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP1 (Strong) is: "Missense or in-frame variant outside critical domains without splicing impact." The evidence for this variant shows: it is a nonsense variant. Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP2 is: "Observed in trans with a pathogenic variant for a fully penetrant dominant disorder." The evidence for this variant shows: no trans observations reported. Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP3 is: "In-frame deletions/insertions in repetitive regions without known function." The evidence for this variant shows: it is a nonsense variant. Therefore, this criterion is not applied.
BP4
BP4 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP4 (Supporting) is: "Computational evidence supports no impact (BayesDel≤0.15 and SpliceAI≤0.1)." The evidence for this variant shows: not applicable for truncating changes. Therefore, this criterion is not applied.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP5 is: "Variant found in a case with an alternate molecular basis for disease." The evidence for this variant shows: no such co-occurrence data. Therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP6 is: "Reputable source reports benign." The evidence for this variant shows: no reputable benign assertions. Therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to VCEP guidelines, the rule for BP7 is: "Synonymous or intronic variant without splicing impact." The evidence for this variant shows: it is a nonsense change. Therefore, this criterion is not applied.

