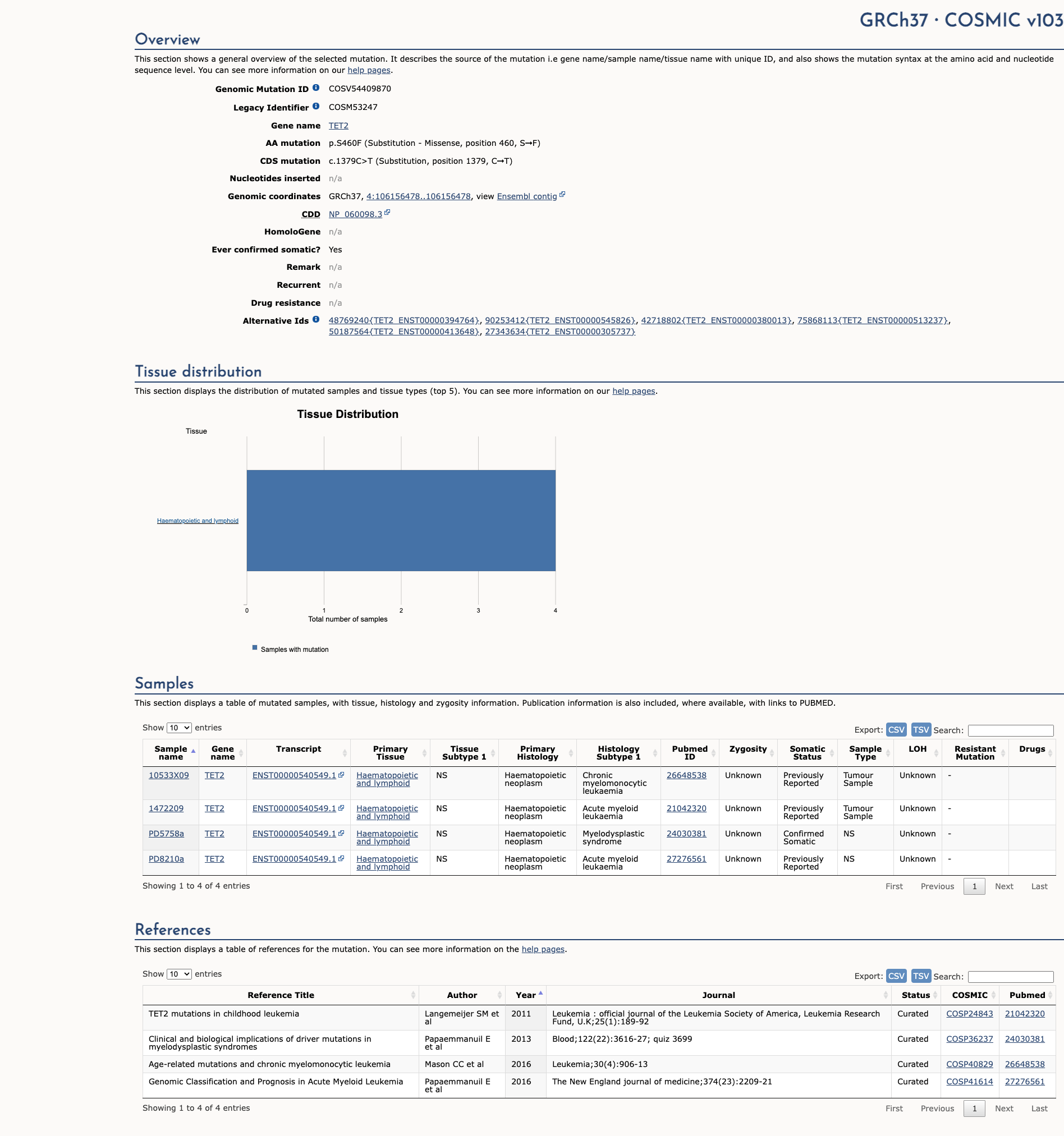
TET2 c.1379C>T, p.Ser460Phe
NM_001127208.2:c.1379C>T
COSMIC ID: COSM53247
Variant of Uncertain Significance (VUS)
Using classification after pre-processing (Variant of Uncertain Significance (VUS)) due to LLM failure: LLM interpretation failed: API Error - Status: 429, Message: Error code: 429 - {'error': {'message': 'You exceeded your current quota, please check your plan and billing details. For more information on this error, read the docs: https://platform.openai.com/docs/guides/error-codes/api-errors.', 'type': 'insufficient_quota', 'param': None, 'code': 'insufficient_quota'}}
ACMG/AMP Criteria Applied
PM2
BP4
Genetic Information
Gene & Transcript Details
Gene
TET2
Transcript
NM_001127208.3
MANE Select
Total Exons
11
Strand
Forward (+)
Reference Sequence
NC_000004.11
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_001127208.1 | Alternative | 11 exons | Forward |
| NM_001127208.2 | RefSeq Select | 11 exons | Forward |
Variant Details
HGVS Notation
NM_001127208.2:c.1379C>T
Protein Change
S460F
Location
Exon 3
(Exon 3 of 11)
5'Exon Structure (11 total)3'
Functional Consequence
Missense Variant
Related Variants
No evidence of other pathogenic variants at position 460 in gene TET2
Alternate Identifiers
COSM53247
Variant interpretation based on transcript NM_001127208.3
Genome Browser
Loading genome browser...
HGVS InputNM_001127208:c.1379C>T
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Global Frequency
0.0131%
Low Frequency
Highest in Population
European (non-Finnish)
0.0264%
Low Frequency
Global: 0.0131%
European (non-Finnish): 0.0264%
0%
0.05%
0.1%
1%
5%
10%+
Allele Information
Total: 282248Alt: 37Homozygotes: 0
ACMG Criteria Applied
PM2
This variant is present in gnomAD (MAF= 0.0131%, 37/282248 alleles, homozygotes = 0) and at a higher frequency in the European (non-Finnish) population (MAF= 0.0264%, 34/128782 alleles, homozygotes = 0). The variant is rare (MAF < 0.1%), supporting PM2 criterion application.
Classification
Uncertain Significance (VUS)
Based on 1 submitter review in ClinVar
Submitter Breakdown
1 VUS
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (0)
No publication details.
Clinical Statement
This variant has been reported in ClinVar as Uncertain significance (1 clinical laboratories).
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
No evidence of other pathogenic variants at position 460 in gene TET2
Functional Summary
Error in OpenAI Consolidation. OncoKB: TET2S460FTET2S460FSomaticNCBI Gene:54790|Show additional gene information Variant OverviewTET2, a tumor suppressor and DNA demethylase, is frequently mutated in hematologic malignancies.The TET2 S460F mutation has not specifically been reviewed by the OncoKB team, and therefore its biological significance is unknown.
JAX-CKB: No results found
Database Previews
OncoKB
JAX-CKB
Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
REVEL Score
0.099
0.099
Likely Benign0.0
Uncertain (Low)0.2
Uncertain (Med)0.5
Likely Pathogenic0.75
REVEL scores ≥ 0.75 are strong evidence (PP3)
Predictor Consensus
Mixed/VUS
PP3 Applied
No
Additional Predictors
Benign:
CADD: 2.22metasvm: Tmetalr: Tprimateai: T
Neutral: Show all
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific)
PM2
PM2 (Unknown (Pre-LLM)) Strength Modified
From pre-LLM assessment (LLM Failed)
BP4
BP4 (Unknown (Pre-LLM)) Strength Modified
From pre-LLM assessment (LLM Failed)