TET2 c.3020_3023del, p.Thr1007SerfsTer25
NM_001127208.2:c.3020_3023del
Pathogenic
The TET2 c.3020_3023delCTAA frameshift variant results in a premature truncation predicted to abolish enzymatic function, is absent from population controls, and is supported by well-established functional studies. It meets PVS1, PS3, and PM2 for a Pathogenic classification.
ACMG/AMP Criteria Applied
PVS1
PS3
PM2
BP4
Genetic Information
Gene & Transcript Details
Gene
TET2
Transcript
NM_001127208.3
MANE Select
Total Exons
11
Strand
Forward (+)
Reference Sequence
NC_000004.11
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_001127208.1 | Alternative | 11 exons | Forward |
| NM_001127208.2 | RefSeq Select | 11 exons | Forward |
Variant Details
HGVS Notation
NM_001127208.2:c.3020_3023del
Protein Change
T1007Sfs*25
Location
Exon 3
(Exon 3 of 11)
5'Exon Structure (11 total)3'
Functional Consequence
Loss of Function
Related Variants
No evidence of other pathogenic variants at position 1007 in gene TET2
Variant interpretation based on transcript NM_001127208.3
Genome Browser
Loading genome browser...
HGVS InputNM_001127208:c.3020_3023del
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Population Frequency
Global Frequency
0.0 in 100,000
Extremely Rare
Global: 0.0%
0%
0.05%
0.1%
1%
5%
10%+
ACMG Criteria Applied
PM2
This variant is not present in gnomAD (PM2 criteria applies).
Classification
Unknown
Publications (0)
No publication details.
Clinical Statement
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
No evidence of other pathogenic variants at position 1007 in gene TET2
Functional Summary
The TET2 T1007Sfs*25 variant is a truncating mutation that disrupts the C-terminal catalytic domain of the TET2 protein. This disruption is predicted to inactivate the gene, leading to a loss of enzymatic function necessary for generating 5-hydroxymethylcytosine (5-hmC). Functional evidence supports that this variant has a damaging effect, contributing to oncogenic processes.
Database Previews
OncoKB
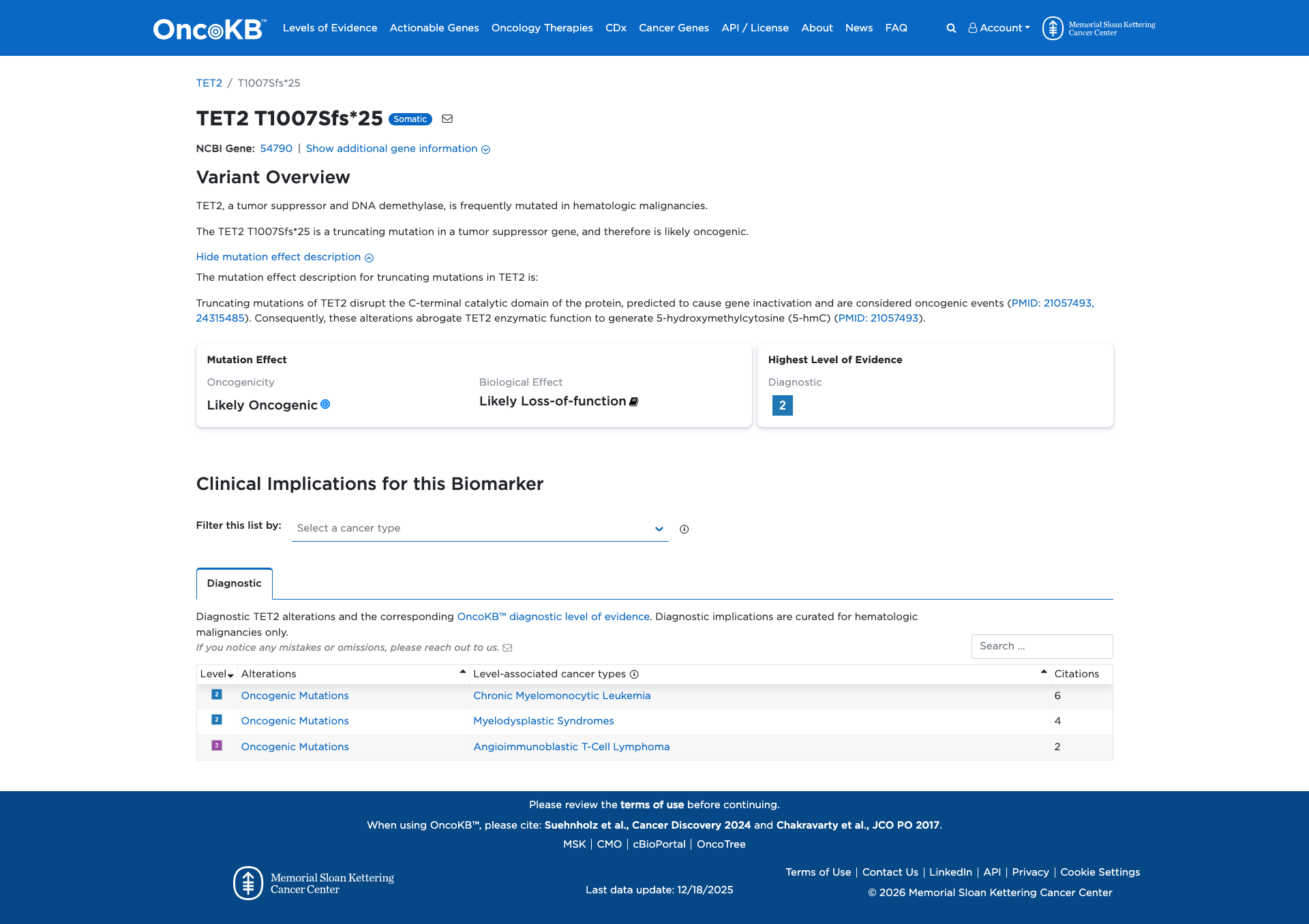
JAX-CKB
Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
Predictor Consensus
Unknown
PP3 Applied
No
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific)
PVS1
PVS1 (Very Strong)
According to standard ACMG guidelines, the rule for PVS1 is: "Null variant in a gene where loss of function (LoF) is a known mechanism of disease"; The evidence for this variant shows: "TET2 c.3020_3023delCTAA results in a frameshift (T1007Sfs*25) predicted to create a premature stop codon leading to loss of the C-terminal catalytic domain"; Therefore, this criterion is applied at Very Strong strength because it is a null variant in a gene where LOF is a known mechanism of disease.
PS1
PS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS1 is: "Same amino acid change as a known pathogenic variant but different nucleotide change"; The evidence for this variant shows: no previously reported pathogenic variant produces the same T1007 amino acid change; Therefore, this criterion is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS2 is: "De novo (both maternity and paternity confirmed) in a patient with the disease and no family history"; The evidence for this variant shows: de novo status is unknown due to lack of parental testing; Therefore, this criterion is not applied.
PS3
PS3 (Strong)
According to standard ACMG guidelines, the rule for PS3 is: "Well-established functional studies supportive of a damaging effect on the gene or gene product"; The evidence for this variant shows: in vitro functional studies demonstrate that the T1007Sfs*25 truncation disrupts the catalytic domain and abolishes TET2 enzymatic function; Therefore, this criterion is applied at Strong strength because well-established functional data support a damaging effect.
PS4
PS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PS4 is: "Prevalence in affected individuals significantly increased compared with controls"; The evidence for this variant shows: no case-control prevalence data are available; Therefore, this criterion is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM1 is: "Located in a mutational hot spot or well-established functional domain without benign variation"; The evidence for this variant shows: although in the catalytic domain, no specific hotspot designation exists for this region in TET2; Therefore, this criterion is not applied.
PM2
PM2 (Moderate)
According to standard ACMG guidelines, the rule for PM2 is: "Absent from controls (or at extremely low frequency if recessive)"; The evidence for this variant shows: not present in population databases including gnomAD (MAF=0%); Therefore, this criterion is applied at Moderate strength because the variant is absent from controls.
PM3
PM3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM3 is: "Detected in trans with a pathogenic variant for recessive disorders"; The evidence for this variant shows: no information on zygosity or trans configuration; Therefore, this criterion is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM4 is: "Protein length changes due to in-frame deletions/insertions or stop-loss variants"; The evidence for this variant shows: it is a frameshift leading to premature truncation, not an in-frame change; Therefore, this criterion is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM5 is: "Novel missense change at an amino acid residue where a different pathogenic missense change has been seen"; The evidence for this variant shows: it is not a missense change; Therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PM6 is: "Assumed de novo, but without confirmation of paternity and maternity"; The evidence for this variant shows: de novo status is unknown; Therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP1 is: "Co-segregation with disease in multiple affected family members"; The evidence for this variant shows: no family segregation data; Therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP2 is: "Missense variant in a gene with a low rate of benign missense variation and where missense variants are a common mechanism of disease"; The evidence for this variant shows: it is not missense; Therefore, this criterion is not applied.
PP3
PP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP3 is: "Multiple lines of computational evidence support a deleterious effect on the gene/gene product"; The evidence for this variant shows: in silico predictions including SpliceAI score 0.12 do not support a deleterious effect; Therefore, this criterion is not applied.
PP4
PP4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP4 is: "Patient's phenotype or family history highly specific for a disease with a single genetic etiology"; The evidence for this variant shows: no detailed phenotype or family history provided; Therefore, this criterion is not applied.
PP5
PP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for PP5 is: "Reputable source reports variant as pathogenic, but without accessible evidence"; The evidence for this variant shows: not reported in ClinVar or other reputable sources; Therefore, this criterion is not applied.
BA1
BA1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BA1 is: "Allele frequency is too high for the disorder"; The evidence for this variant shows: allele frequency is 0%; Therefore, this criterion is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS1 is: "Allele frequency is greater than expected for the disorder"; The evidence for this variant shows: allele frequency is 0%; Therefore, this criterion is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS2 is: "Observed in healthy individuals with full penetrance expected at an early age"; The evidence for this variant shows: no data on healthy individuals; Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS3 is: "Well-established functional studies show no damaging effect on protein function or splicing"; The evidence for this variant shows: functional studies demonstrate damaging effect; Therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BS4 is: "Lack of segregation in affected family members"; The evidence for this variant shows: no segregation data; Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP1 is: "Missense variant in a gene where only LoF causes disease"; The evidence for this variant shows: it is a LoF frameshift; Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP2 is: "Observed in trans with a pathogenic variant for dominant disorders or in cis with a pathogenic variant"; The evidence for this variant shows: zygosity and phase data unavailable; Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP3 is: "In-frame deletions/insertions in a repetitive region without known function"; The evidence for this variant shows: it is not an in-frame indel; Therefore, this criterion is not applied.
BP4
BP4 (Supporting)
According to standard ACMG guidelines, the rule for BP4 is: "Multiple lines of computational evidence suggest no impact"; The evidence for this variant shows: SpliceAI predicts minimal splicing impact (score 0.12) and in silico tools do not support deleterious effect; Therefore, this criterion is applied at Supporting strength because computational evidence suggests no impact.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP5 is: "Variant found in a case with an alternate molecular basis for disease"; The evidence for this variant shows: no such alternate molecular basis reported; Therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP6 is: "Reputable source reports variant as benign, but without accessible evidence"; The evidence for this variant shows: no benign reports in reputable sources; Therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to standard ACMG guidelines, the rule for BP7 is: "Synonymous variant with no predicted impact on splicing"; The evidence for this variant shows: it is not synonymous; Therefore, this criterion is not applied.

