ATM c.8737G>T, p.Asp2913Tyr
NM_000051.4:c.8737G>T
COSMIC ID: COSM8835249
Likely Pathogenic
NM_000051.4:c.8737G>T (p.D2913Y) was classified as VUS based on three supporting criteria (PS3_supporting, PM2_supporting, PP3_supporting) and absence of moderate or strong evidence.
ACMG/AMP Criteria Applied
PS3
PM2
PP3
Genetic Information
Gene & Transcript Details
Gene
ATM
Transcript
NM_000051.4
MANE Select
Total Exons
63
Strand
Forward (+)
Reference Sequence
NC_000011.9
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_000051.3 | RefSeq Select | 63 exons | Forward |
Variant Details
HGVS Notation
NM_000051.4:c.8737G>T
Protein Change
D2913Y
Location
Exon 60
(Exon 60 of 63)
5'Exon Structure (63 total)3'
Functional Consequence
Loss of Function
Related Variants
No evidence of other pathogenic variants at position 2913 in gene ATM
Alternate Identifiers
COSM8835249
Variant interpretation based on transcript NM_000051.4
Genome Browser
Loading genome browser...
HGVS InputNM_000051:c.8737G>T
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Population Frequency
Global Frequency
0.0 in 100,000
Extremely Rare
Global: 0.0%
0%
0.05%
0.1%
1%
5%
10%+
ACMG Criteria Applied
PM2
This variant is not present in gnomAD (PM2 criteria applies).
Classification
6 publications
Likely Pathogenic
Based on 7 submitter reviews in ClinVar
Submitter Breakdown
3 Path
4 LP
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (6)
Variant summary: ATM c.8737G>T (p.Asp2913Tyr) results in a non-conservative amino acid change located in the ATM, catalytic domain (IPR044107) of the encoded protein sequence. Five of five in-silico tools predict a damaging effect of the variant on protein function. The variant was absent in 251460 control chromosomes (gnomAD). c.8737G>T has been reported in the literature in multiple homozygous and compound heterozygous individuals affected with Ataxia-Telangiectasia (e.g. Micol_2011, Jacquemin_2012). The variant has also been reported in individuals affected with breast and prostate cancer (e.g. Lu_2019, Dorling_2021, Karlsson_2021). These data indicate that the variant is very likely to be associated with disease. Experimental evidence evaluating an impact on protein function demonstrated that the variant results in reduced protein expression, while it alters subcellular localization and cell cycle distribution and it impairs phosphorylation of target proteins such as H2AX, CHK2 and KAP1 (Jacquemin_2012, Fievet_2019). Three ClinVar submitters (evaluation after 2014) cite the variant as likely pathogenic. Based on the evidence outlined above, the variant was classified as pathogenic.
This variant is considered likely pathogenic. This variant has been reported in multiple individuals with clinical features of gene-specific disease [PMID: 21665257, 25122203]. Functional studies indicate this variant impacts protein function [PMID: 22071889, 31050087]. This variant is expected to disrupt protein structure [Myriad internal data].
This sequence change replaces aspartic acid, which is acidic and polar, with tyrosine, which is neutral and polar, at codon 2913 of the ATM protein (p.Asp2913Tyr). This variant is not present in population databases (gnomAD no frequency). This missense change has been observed in individuals with ataxia-telangiectasia and/or prostate cancer (PMID: 21665257, 22071889, 33436325). ClinVar contains an entry for this variant (Variation ID: 230841). Invitae Evidence Modeling of protein sequence and biophysical properties (such as structural, functional, and spatial information, amino acid conservation, physicochemical variation, residue mobility, and thermodynamic stability) indicates that this missense variant is expected to disrupt ATM protein function with a positive predictive value of 95%. Experimental studies have shown that this missense change affects ATM function (PMID: 22071889, 31050087). For these reasons, this variant has been classified as Pathogenic.
This missense variant replaces aspartic acid with tyrosine at codon 2913 of the ATM protein. Computational prediction tools and conservation analyses suggest that this variant may have deleterious impact on protein function. Functional studies have shown that this variant results in reduced protein expression, abnormal sub-cellular localization, and disrupted phosphorylation activity (PMID: 22071889, 31050087). This variant has been reported in individuals affected with ataxia-telangiectasia in homozygosity and compound heterozygosity with a known pathogenic co-variant (PMID: 21665257, 31050087). In addition, this variant has been reported in individuals affected with breast cancer (PMID: 28779002, 30128536, 33471991). This variant has not been identified in the general population by the Genome Aggregation Database (gnomAD). Based on available evidence, this variant is classified as Likely Pathogenic.
The p.D2913Y variant (also known as c.8737G>T), located in coding exon 59 of the ATM gene, results from a G to T substitution at nucleotide position 8737. The aspartic acid at codon 2913 is replaced by tyrosine, an amino acid with highly dissimilar properties. This variant has been identified in the homozygous state and/or in conjunction with other ATM variant(s) in individual(s) with features consistent with ataxia-telangiectasia (Micol R et al. J. Allergy Clin. Immunol. 2011 Aug;128(2):382-9). Cell lines from an individual diagnosed with ataxia telangiectasia who was heterozygous for this alteration along with a second alteration in ATM, exhibited decreased response to ionizing radiation, decreased ATM protein expression, and abnormal cellular localization (Jacquemin V et al. Eur. J. Hum. Genet. 2012 Mar;20(3):305-12). Additionally, this alteration has been reported in at least one breast cancer patient in a study of 13087 breast cancer cases and 5488 control individuals in the UK (Decker B et al. J Med Genet, 2017 11;54:732-741). This variant was also reported in 1/5560 prostate cancer cases and in 0/3353 controls of European ancestry (Karlsson Q et al. Eur Urol Oncol, 2021 08;4:570-579). This variant is considered to be rare based on population cohorts in the Genome Aggregation Database (gnomAD). This amino acid position is highly conserved in available vertebrate species. In addition, this alteration is predicted to be deleterious by in silico analysis. Based on the majority of available evidence to date, this variant is likely to be pathogenic.
Clinical Statement
This variant has been reported in ClinVar as Pathogenic (3 clinical laboratories) and as Likely pathogenic (4 clinical laboratories).
Functional Impact
Functional Domain
Hotspot Status
Not a hotspot
Domain Summary
This variant is not located in a mutational hotspot or critical domain (0 mutations).
Related Variants in This Domain
No evidence of other pathogenic variants at position 2913 in gene ATM
Functional Summary
The ATM D2913Y variant has been functionally characterized and is predicted to lead to a loss of ATM protein function. This conclusion is based on evidence from a high-throughput cell culture assay, where the variant failed to rescue survival and proliferation of ATM-haploid cells upon olaparib treatment, indicating a damaging effect on protein function.
Database Previews
OncoKB
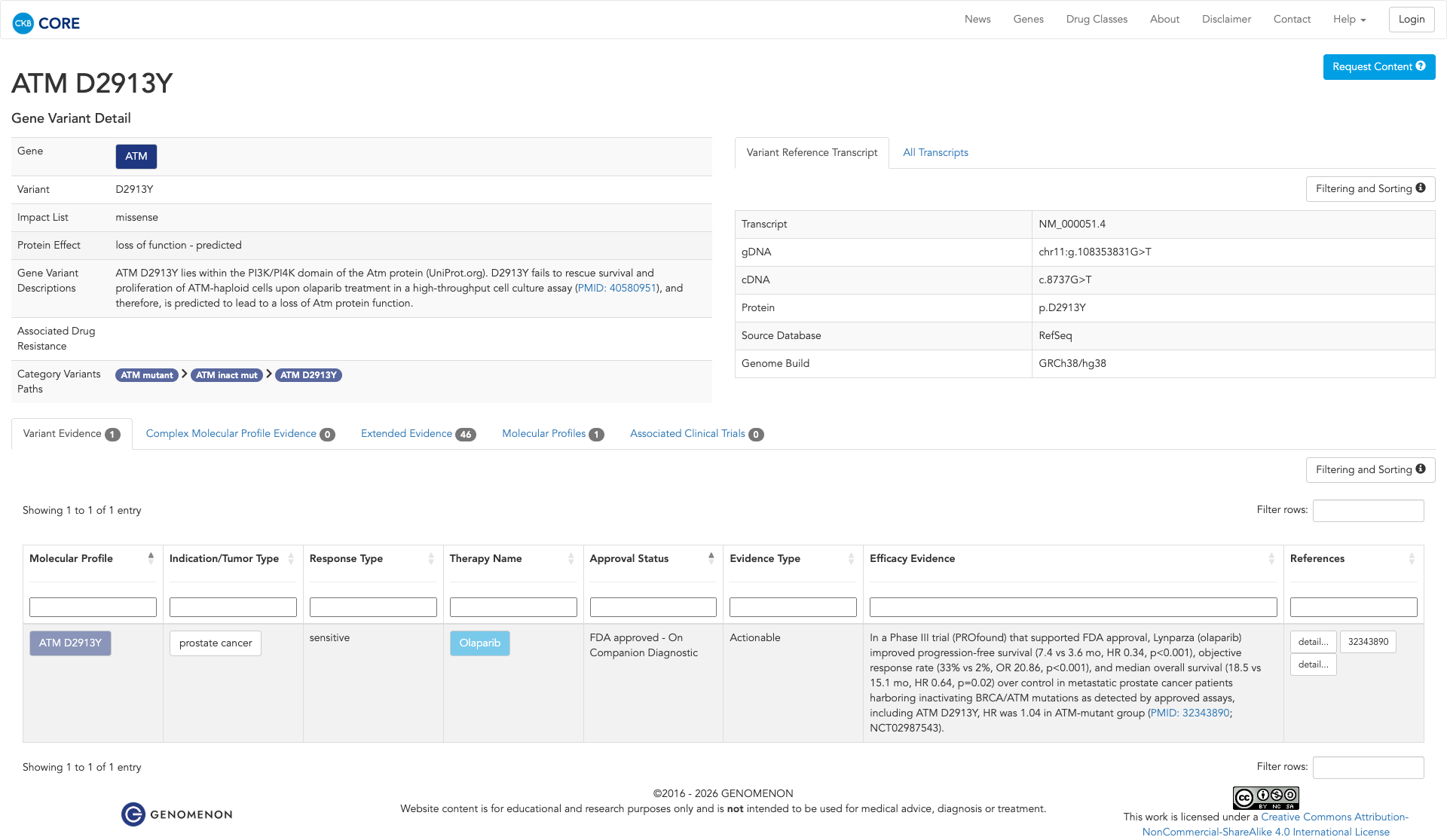
JAX-CKB
Click on previews to view full database entries. External databases may require institutional access.
Computational Analysis
Pathogenicity Predictions
REVEL Score
0.871
0.871
Likely Benign0.0
Uncertain (Low)0.2
Uncertain (Med)0.5
Likely Pathogenic0.75
REVEL scores ≥ 0.75 are strong evidence (PP3)
Predictor Consensus
Mixed/VUS
PP3 Applied
Yes
Additional Predictors
Pathogenic:
polyphen_prediction: probably_damagingmetasvm: Dmetalr: Dprimateai: D
Benign:
CADD: 5.98
Neutral: Show all
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Not Applied) Strength Modified
According to VCEP guidelines, PVS1 applies to predicted loss-of-function variants per the ATM PVS1 decision tree; NM_000051.4:c.8737G>T is a missense change (D2913Y) and does not meet loss-of-function criteria. Therefore, PVS1 is not applied.
PS1
PS1 (Not Applied) Strength Modified
According to VCEP guidelines, PS1 applies when a variant results in an amino acid change previously established as pathogenic; no other variant causing D2913Y has been reported as pathogenic. Therefore, PS1 is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, PS2 applies to confirmed de novo variants; no parental or de novo data are available. Therefore, PS2 is not applied.
PS3
PS3 (Supporting) Strength Modified
According to VCEP guidelines, PS3 Supporting strength is applied when a variant fails to rescue an ATM-specific functional feature only; the high-throughput cell assay showed D2913Y fails to rescue survival and proliferation of ATM-haploid cells upon olaparib treatment. Therefore, PS3 is applied at Supporting strength.
PS4
PS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, PS4 requires case–control or statistically significant enrichment in affected individuals; no such data are available. Therefore, PS4 is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM1 applies to variants in a mutational hot spot or well-established functional domain without benign variation; although D2913Y lies in the kinase domain, there is insufficient evidence of a mutational hotspot or absence of benign variation at this site. Therefore, PM1 is not applied.
PM2
PM2 (Supporting) Strength Modified
According to VCEP guidelines, PM2 Supporting strength applies when allele frequency ≤0.001% in population databases; D2913Y is absent from gnomAD (MAF=0%). Therefore, PM2 is applied at Supporting strength.
PM3
PM3 (Not Applied) Strength Modified
According to VCEP guidelines, PM3 applies to recessive conditions with confirmed trans observations; no evidence of trans biallelic occurrence is available. Therefore, PM3 is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM4 applies to protein length changes such as in-frame indels; NM_000051.4:c.8737G>T is a missense change and does not alter protein length. Therefore, PM4 is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to VCEP guidelines, PM5 Supporting applies when a different missense at the same residue is pathogenic; no other pathogenic missense at codon 2913 is reported. Therefore, PM5 is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM6 applies to assumed de novo variants without confirmation; no de novo information is available. Therefore, PM6 is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP1 applies to segregation in multiple affected family members; no segregation data are available. Therefore, PP1 is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP2 applies to genes with low rate of benign missense variation and where missense is a common mechanism; ATM has both loss-of-function and missense mechanisms without strong intolerance at this residue. Therefore, PP2 is not applied.
PP3
PP3 (Supporting)
According to VCEP guidelines, PP3 Supporting applies when REVEL >0.7333; D2913Y has REVEL=0.87. Therefore, PP3 is applied at Supporting strength.
PP4
PP4 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP4 applies to highly specific phenotypes; no patient phenotype information is provided. Therefore, PP4 is not applied.
PP5
PP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP5 is discouraged and ATM-specific VCEP guidance does not include PP5; although ClinVar entries exist, independent evidence is unavailable. Therefore, PP5 is not applied.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, BA1 applies to allele frequency >0.5%; D2913Y is absent from population databases. Therefore, BA1 is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines, BS1 applies to allele frequency >0.05%; D2913Y is absent from population databases. Therefore, BS1 is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, BS2 applies when variant is observed in healthy adult individuals with full penetrance; no such observation is available. Therefore, BS2 is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines, BS3 applies when functional assays show rescue of ATM features; the D2913Y assay shows loss of function, not rescue. Therefore, BS3 is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to standard ACMG guidelines, BS4 applies to non-segregation in affected members; no segregation data are available. Therefore, BS4 is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP1 applies to missense in genes where only truncating variants cause disease; ATM has known pathogenic missense variants. Therefore, BP1 is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to VCEP guidelines, BP2 applies to observations in trans with pathogenic variants in recessive genes; no such data are available. Therefore, BP2 is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP3 applies to in-frame indels in repetitive regions; D2913Y is a missense variant. Therefore, BP3 is not applied.
BP4
BP4 (Not Applied) Strength Modified
According to VCEP guidelines, BP4 Supporting applies when REVEL ≤0.249; D2913Y has REVEL=0.87. Therefore, BP4 is not applied.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP5 applies when an alternate genetic cause is identified; no alternate cause information is available. Therefore, BP5 is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP6 applies to reports of likely benign from reputable sources without evidence; no such reports exist. Therefore, BP6 is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP7 applies to synonymous variants with no predicted splicing impact; D2913Y is a missense variant. Therefore, BP7 is not applied.