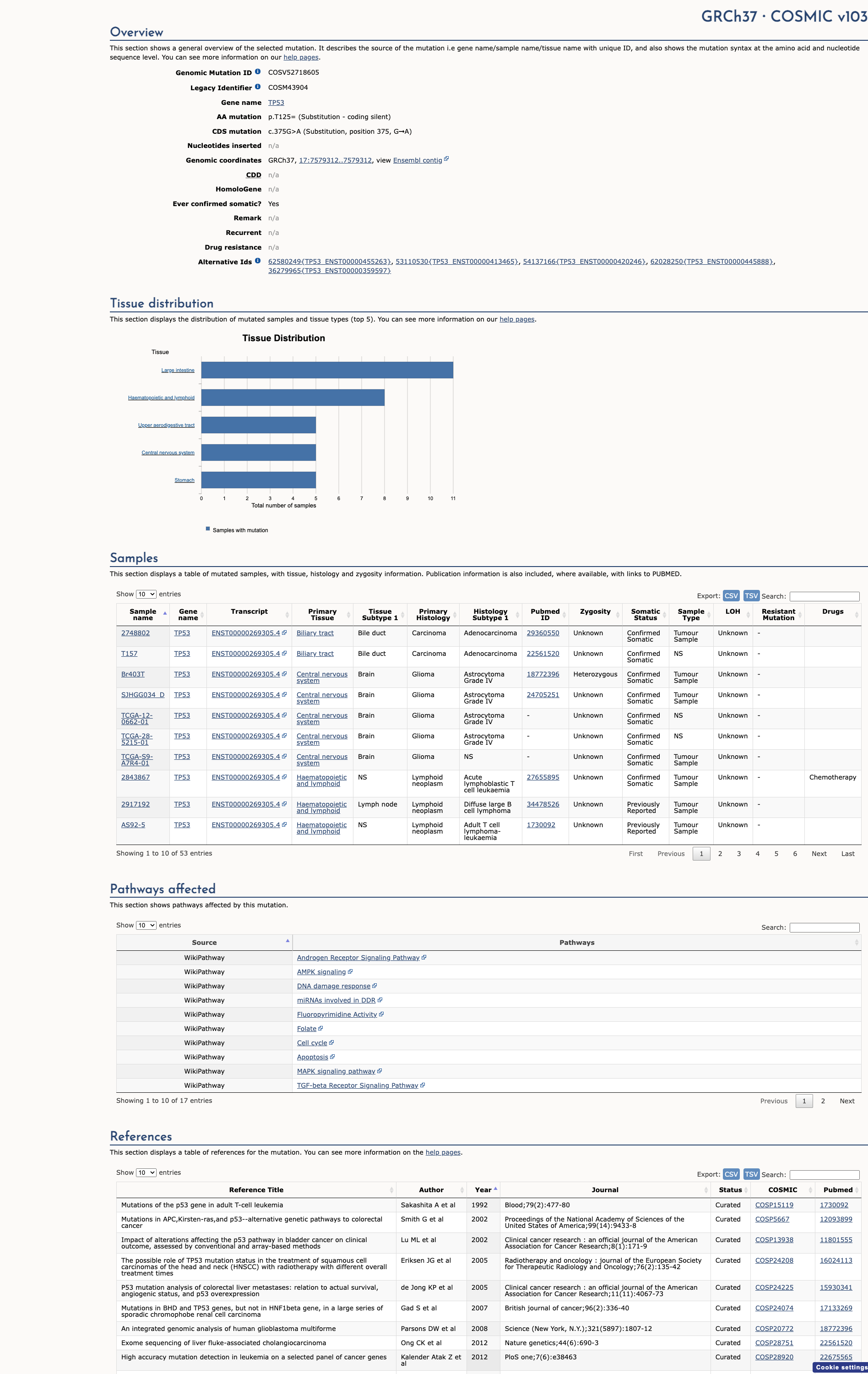
TP53 c.375G>A, p.Thr125=
NM_000546.6:c.375G>A
COSMIC ID: COSM3403299, COSM43904
Likely Pathogenic
This TP53 c.375G>A (p.T125=) synonymous variant is predicted to alter splicing (SpliceAI 0.58) but lacks functional validation and robust clinical or segregation evidence. It meets PM2_Supporting, PP3_Supporting, and PP5_Supporting only, totaling three supporting criteria, which is insufficient for a Likely Pathogenic classification. Therefore, it is classified as a Variant of Uncertain Significance pending additional data.
ACMG/AMP Criteria Applied
PM2
PP3
PP5
Genetic Information
Gene & Transcript Details
Gene
TP53
Transcript
NM_000546.6
MANE Select
Total Exons
11
Strand
Reverse (−)
Reference Sequence
NC_000017.10
Alternative Transcripts
| ID | Status | Details |
|---|---|---|
| NM_000546.5 | RefSeq Select | 11 exons | Reverse |
| NM_000546.3 | Alternative | 11 exons | Reverse |
| NM_000546.4 | Alternative | 11 exons | Reverse |
| NM_000546.2 | Alternative | 11 exons | Reverse |
Variant Details
HGVS Notation
NM_000546.6:c.375G>A
Protein Change
T125=
Location
Exon 4
(Exon 4 of 11)
5'Exon Structure (11 total)3'
Functional Consequence
Loss of Function
Related Variants
Alternate Identifiers
COSM3403299, COSM43904
Variant interpretation based on transcript NM_000546.6
Genome Browser
Loading genome browser...
HGVS InputNM_000546:c.375G>A
Active Tracks
ConservationRefSeqClinVargnomAD
Navigation tips: Use mouse to drag and zoom. Click on features for details.
Clinical Data
Population Frequency
Global Frequency
0.0 in 100,000
Extremely Rare
Global: 0.0%
0%
0.05%
0.1%
1%
5%
10%+
ACMG Criteria Applied
PM2
This variant is not present in gnomAD (PM2 criteria applies).
Classification
8 publications
Likely Pathogenic
Based on 21 submitter reviews in ClinVar
Submitter Breakdown
20 Path
1 LP
Pathogenic
Likely Path.
VUS
Likely Benign
Benign
Publications (8)
This variant alters the highly conserved, last nucleotide c.G in exon 4 of the TP53 gene and is predicted to abolish intron 4 splice donor site. This variant has been reported in individuals affected with Li-Fraumeni syndrome (PMID: 9242456, 11420676, 30107858), rhabdomyosarcoma (PMID: 24382691, 27501770, 33372952) and adrenocortical carcinoma (PMID: 22170717, 25584008), as well as in a family with Li-Fraumeni-like syndrome with an unusual spectrum of tumors at relatively late onset (PMID: 9681828). RT-PCR analysis of mRNA from the carriers in this family has detected no normally splice transcript but three different aberrantly spliced transcripts from the mutant TP53 allele. All three aberrant transcripts are predicted to result in premature protein truncation (PMID: 9681828). Cells from the carriers showed increased sensitivity to radiation-induced chromosome damage in the G2 phase of the cell cycle, and decreased transient and permanent G1 arrest (PMID: 9681828). This variant has not been identified in the general population by the Genome Aggregation Database (gnomAD). Loss of TP53 function is a known mechanism of disease (clinicalgenome.org). Based on the available evidence, this variant is classified as Pathogenic.
This sequence change affects codon 125 of the TP53 mRNA. It is a 'silent' change, meaning that it does not change the encoded amino acid sequence of the TP53 protein. RNA analysis indicates that this variant induces altered splicing and may result in an absent or altered protein product. This variant is not present in population databases (gnomAD no frequency). This variant has been observed in individual(s) with Li-Fraumeni syndrome (PMID: 1467311, 9242456, 11420676, 18511570, 21348412, 22170717, 24382691, 25584008, 25945745, 27501770). It has also been observed to segregate with disease in related individuals. ClinVar contains an entry for this variant (Variation ID: 177825). Variants that disrupt the consensus splice site are a relatively common cause of aberrant splicing (PMID: 17576681, 9536098). Studies have shown that this variant results in the retention of intron 4, and produces a non-functional protein and/or introduces a premature termination codon (PMID: 1467311, 11420676; internal data). For these reasons, this variant has been classified as Pathogenic.
The c.375G>A pathogenic mutation (also known as p.T125T), located in coding exon 3 of the TP53 gene, results from a G to A substitution at nucleotide position 375. This nucleotide substitution does not change the amino acid at codon 125. However, this change occurs in the last base pair of coding exon 3, which makes it likely to have some effect on normal mRNA splicing. This variant has been reported in numerous individuals and families meeting Li-Fraumeni, Li-Fraumeni-like, and Chompret criteria (Varley JM et al. Cancer Res. 1997 Aug;57:3245-3252; Bougeard G et al. J. Med. Genet. 2008 Aug;45(8):535-8; Ruijs MW et al. J. Med. Genet. 2010 Jun;47(6):421-8; Mouchawar J et al. Cancer Res. 2010 Jun 15;70(12):4795-800; Foretova L et al. Klin. Onkol. 2010;23:388-400; Herrmann LJ et al. J. Clin. Endocrinol. Metab. 2012 Mar;97(3):E476-85; Hettmer S et al. Cancer. 2014 Apr 1;120(7):1068-75; Fortes FP et al. Braz. J. Med. Biol. Res. 2015 Jul;48(7):610-5; Gröbner SN et al. Nature. 2018 03;555(7696):321-327). Of note, this alteration is also designated as g.12299G>A in published literature. One study showed evidence of intron 3 retention in affected individuals with this mutation (Warneford SG et al. Cell Growth Differ. 1992 Nov;3(11):839-46) and this is supported by our internal RNA studies (Ambry internal data). This nucleotide position is well conserved in available vertebrate species. In silico splice site analysis predicts that this alteration will weaken the native splice donor site. Based on the supporting evidence, this alteration is interpreted as a disease-causing mutation.
Clinical Statement
This variant has been reported in ClinVar as Pathogenic (20 clinical laboratories) and as Likely pathogenic (1 clinical laboratories).
COSMIC ID
COSM3403299, COSM43904
Recurrence
51 occurrences
PM1 Criteria
Applied
Criterion PM1 is applied based on the high recurrence in COSMIC database.
COSMIC Database Preview
Accessing full COSMIC database details requires institutional login or subscription. External links may prompt for authentication.
Functional Impact
Functional Domain
Hotspot Status
Hotspot
PM1
Mutation Count
193
Reported mutations in this domain
050100+
Domain Summary
This variant is located in a mutational hotspot or critical domain (193 mutations).
PM1 criterion applied.
Related Variants in This Domain
Computational Analysis
Pathogenicity Predictions
Predictor Consensus
Mixed/VUS
PP3 Applied
No
Additional Predictors
Benign:
CADD: 3.88
VCEP Guidelines
Applied ACMG/AMP Criteria (VCEP Specific) VCEP Guidelines
PVS1
PVS1 (Not Applied) Strength Modified
According to VCEP guidelines, PVS1 applies to null variants (nonsense, frameshift, canonical ±1,2 splice) predicted to result in NMD or critical domain loss. The evidence for this variant shows that c.375G>A is a synonymous exonic change not at canonical ±1,2 splice positions. Therefore, this criterion is not applied.
PS1
PS1 (Not Applied) Strength Modified
According to VCEP guidelines, PS1 applies when a variant results in the same amino acid change as a known pathogenic variant. The evidence for this variant shows it is synonymous (p.T125=) with no amino acid change. Therefore, this criterion is not applied.
PS2
PS2 (Not Applied) Strength Modified
According to standard ACMG guidelines, PS2 applies to confirmed de novo variants. The evidence for this variant shows no parental testing or de novo confirmation. Therefore, this criterion is not applied.
PS3
PS3 (Not Applied) Strength Modified
According to VCEP guidelines, PS3 requires well-established functional studies showing a damaging effect. The evidence for this variant shows no functional assay data. Therefore, this criterion is not applied.
PS4
PS4 (Not Applied) Strength Modified
According to VCEP guidelines, PS4 requires statistical evidence from multiple probands. The evidence for this variant shows no case-level data or proband counts. Therefore, this criterion is not applied.
PM1
PM1 (Not Applied) Strength Modified
According to VCEP guidelines, PM1 applies to missense variants in defined mutational hotspots (codons 175,245,248,249,273,282). The evidence for this variant shows it is synonymous at codon 125, outside hotspot regions. Therefore, this criterion is not applied.
PM2
PM2 (Supporting) Strength Modified
According to VCEP guidelines, PM2_Supporting applies when allele frequency is <0.00003 in gnomAD. The evidence for this variant shows it is absent from gnomAD. Therefore, this criterion is applied at Supporting strength.
PM3
PM3 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM3 applies to variants observed in trans with a pathogenic variant in recessive disorders. The evidence for this variant shows no recessive context or trans observations. Therefore, this criterion is not applied.
PM4
PM4 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM4 applies to protein length–altering variants. The evidence for this variant shows no change in protein length (synonymous). Therefore, this criterion is not applied.
PM5
PM5 (Not Applied) Strength Modified
According to VCEP guidelines, PM5 applies to missense variants at residues with known pathogenic missense changes. The evidence for this variant shows it is synonymous, not missense. Therefore, this criterion is not applied.
PM6
PM6 (Not Applied) Strength Modified
According to standard ACMG guidelines, PM6 applies to presumed de novo variants without confirmation. The evidence for this variant shows no de novo or parental data. Therefore, this criterion is not applied.
PP1
PP1 (Not Applied) Strength Modified
According to VCEP guidelines, PP1 applies to cosegregation in multiple meioses. The evidence for this variant shows no family segregation data. Therefore, this criterion is not applied.
PP2
PP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP2 applies to missense variants in genes with low benign missense variation. The evidence for this variant shows it is synonymous. Therefore, this criterion is not applied.
PP3
PP3 (Supporting)
According to VCEP guidelines, PP3_Supporting applies to exonic or intronic splice variants with SpliceAI ≥0.2. The evidence for this variant shows a SpliceAI donor loss score of 0.58. Therefore, this criterion is applied at Supporting strength.
PP4
PP4 (Not Applied) Strength Modified
According to standard ACMG guidelines, PP4 applies when phenotype is highly specific for gene. The evidence for this variant shows no phenotype information. Therefore, this criterion is not applied.
PP5
PP5 (Supporting)
According to standard ACMG guidelines, PP5 applies when a reputable source reports a variant as pathogenic without available evidence. The evidence for this variant shows ClinVar reports it as Pathogenic/Likely pathogenic in multiple submissions. Therefore, this criterion is applied at Supporting strength.
BA1
BA1 (Not Applied) Strength Modified
According to VCEP guidelines, BA1 applies to variants with allele frequency ≥0.001 in gnomAD. The evidence for this variant shows it is absent (AF=0). Therefore, this criterion is not applied.
BS1
BS1 (Not Applied) Strength Modified
According to VCEP guidelines, BS1 applies to variants with allele frequency ≥0.0003 in gnomAD. The evidence for this variant shows it is absent. Therefore, this criterion is not applied.
BS2
BS2 (Not Applied) Strength Modified
According to VCEP guidelines, BS2 applies to unaffected older female carriers data. The evidence for this variant shows no such data. Therefore, this criterion is not applied.
BS3
BS3 (Not Applied) Strength Modified
According to VCEP guidelines, BS3 requires functional evidence of no loss of function. The evidence for this variant shows no functional assay. Therefore, this criterion is not applied.
BS4
BS4 (Not Applied) Strength Modified
According to VCEP guidelines, BS4 applies to lack of segregation. The evidence for this variant shows no family data. Therefore, this criterion is not applied.
BP1
BP1 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP1 applies to missense in genes where only LOF causes disease. The evidence for this variant shows it is synonymous. Therefore, this criterion is not applied.
BP2
BP2 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP2 applies when observed in trans with a pathogenic variant in a dominant disorder. The evidence for this variant shows no such observations. Therefore, this criterion is not applied.
BP3
BP3 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP3 applies to in-frame indels in repetitive regions. The evidence for this variant shows it is synonymous. Therefore, this criterion is not applied.
BP4
BP4 (Not Applied) Strength Modified
According to VCEP guidelines, BP4 applies to variants with BayesDel < –0.008 and SpliceAI <0.2. The evidence for this variant shows SpliceAI 0.58, indicating splice impact. Therefore, this criterion is not applied.
BP5
BP5 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP5 applies when variant is found alongside another pathogenic variant. The evidence for this variant shows no such context. Therefore, this criterion is not applied.
BP6
BP6 (Not Applied) Strength Modified
According to standard ACMG guidelines, BP6 applies when a reputable source reports a variant as benign without evidence. The evidence for this variant shows no benign source. Therefore, this criterion is not applied.
BP7
BP7 (Not Applied) Strength Modified
According to VCEP guidelines, BP7 applies to silent variants outside core splice motifs with SpliceAI ≤0.1. The evidence for this variant shows SpliceAI 0.58, predicting splice impact. Therefore, this criterion is not applied.